Research
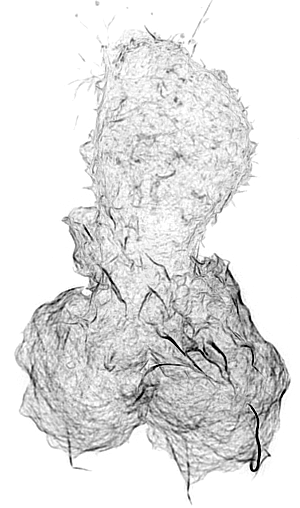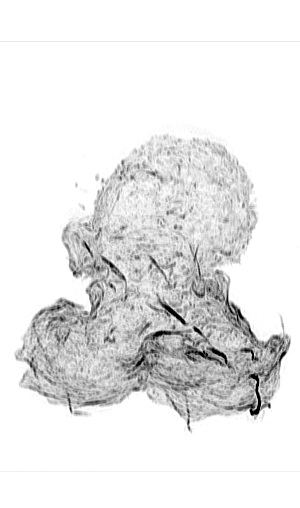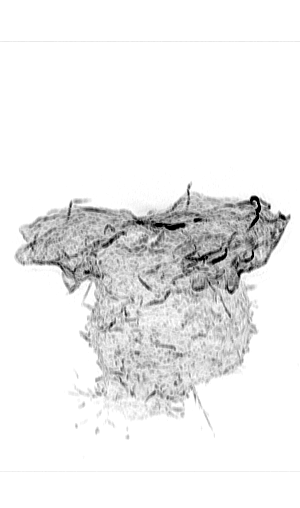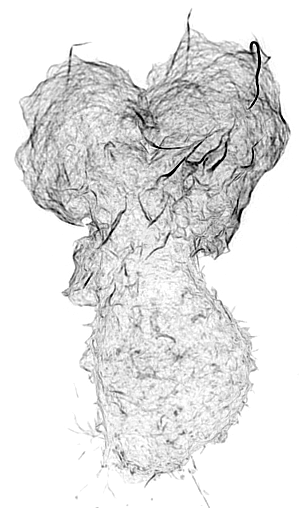
Leukocyte Actin Cytoskeleton
Cellular adhesion and migration require an intricate orchestration of membrane receptors and the cytoskeleton to transduce bidirectional force from extracellular substrates. When dysregulated, a myriad of pathologies can emerge from decreased immune extravasation (leukocyte adhesion deficiency), aberrant immune infiltration (rheumatoid arthritis, psoriasis, inflammatory bowel disease, and atherosclerosis) and tumor metastasis via epithelial-mesenchymal transition. While the importance of the forces generated between cell-cell/ECM and the cytoskeleton are appreciated in cellular regulation, it is unknown how force sensing surface receptors and adhesion complexes dynamically respond to forces and transduce mechanical information. Integrins are one such family of cell surface mechanoreceptors.
Integrin signaling is complex due to their capacity to transmit signals bi-directionally from their extracellular ligand-binding domain, conveying information about substrate availability and stiffness, and from their cytoplasmic domains linking to signal transduction pathways. These intracellular signaling pathways transmit information from chemokine receptors to modulate the activation state of integrins and represent canonical inside-out signaling. Recent evidence supports another branch of inside-out signaling that involves a role for a mechanical component and the actin cytoskeleton. Integrin activation is dependent on their ability to transduce mechanical forces between the actin cytoskeleton and immobilized or cell surface-bound ligand. It is apparent that this force transduction acts as an allosteric effector resulting in integrin conformational change and associated changes in affinity, but the molecular mechanisms driving integrin activation and their regulation at the cellular level remain unclear and is essential to understanding how integrins function to mediate dynamic cellular adhesion and migration.
This cytoskeletal force model of integrin activation inherently links cell polarization and the actin cytoskeleton with integrin activation at the same location within the cell, preventing aberrant integrin-mediated adhesion by limiting active integrins only to where the cell can gain traction. To elucidate the mechanisms of actin’s mechanoregulation of integrin activation will require both functional studies and the development of new methods to measure actin cytoskeleton and integrin dynamics quantitatively. We will utilize super-resolution microscopy, computational image analysis, genome modification, and protein chemistry to understand the cytoskeleton’s linkage to integrins and directly measure integrin conformation on the cell surface, advancing our understanding of the regulation of cell adhesiveness and migration.
