Areas of Interest
Research Interests
Spatial and temporal regulation of gene expression is a key to organismal development. Such precise regulation requires multi-layer of gene expression regulation on transcription, including transcription factor binding, histone modification, DNA modification, and 3D genomic organization. Amazingly, such regulations involve millions of regulatory elements (cis-regulatory elements: enhancers, insulators, etc.) orchestrate on a daily basis to ensure the performance of normal physiological and developmental processes. The dysregulation of such processes could lead to the ectopic gene expression that initiates multiple diseases, particularly cancer. This cis-regulatory elements’ regulation of gene expression still lacks the mechanistic details especially on how transcription is controlled by multi-layer regulation machinery. The Zhang lab aims to understand such multi-layer process on the gene expression regulation in disease and development. We are especially interested in how these gene expression control programs are hijacked in diseases like cancer. Particularly, we are interested in the development of novel small molecule drugs and gene therapies.
The Zhang lab primarily use on the normal and malignant hematopoiesis as models and focus on research of the following directions:
- Transcription hijacking of mutant NPM1 in leukemia.
Mutations in the NPM1 gene are the most common in acute myeloid leukemia (AML). Mutant NPM1’s localization changes from nucleolus to cytoplasm and is highly associated with high HOX gene expression. NPM1 as an intrinsic disordered protein (IDP), study on mutant NPM1’s function in leukemia development and maintenance is murky for long time. We made the seminal discovery of the mutant NPM1 protein as a transcriptional amplifier on chromatin, which transformed the study of this common leukemia mutant protein in acute myeloid leukemia (Fig.1). We are current employing a full spectrum of technologies: epigenome profiling methods including CUT&RUN, RNA immuno-precipitation, protein condensation assay, and super-resolution microscopy to dissect the function and formation of mutant NPM1 on transcription hijacking. These mechanisms now lead us to develop the target therapy for NPM1c AML.
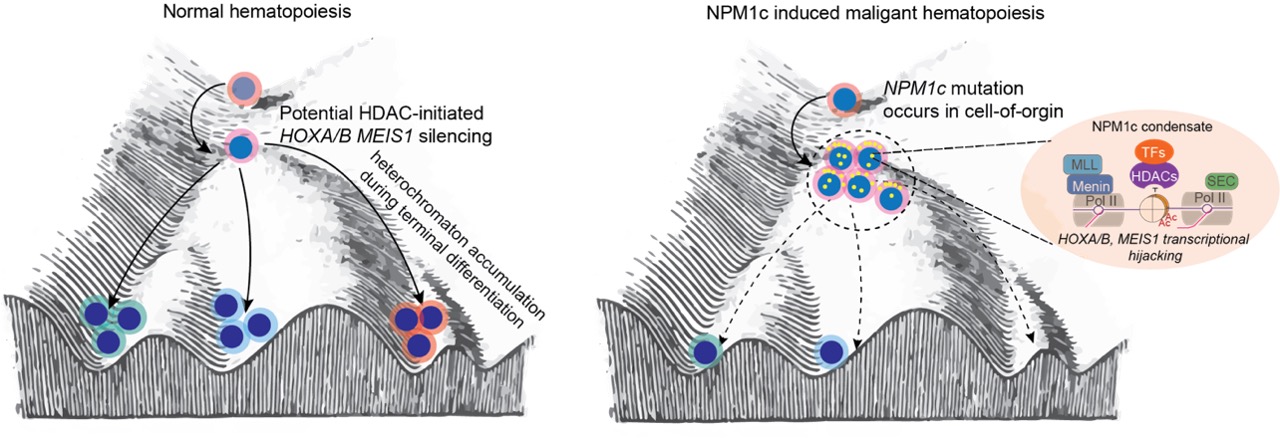
Fig 1. The model of NPM1c transcriptional hijacking during leukemia development
- Function and 3D organization of cis-regulatory elements (CREs)s in gene expression during development.
Enhancer-promoter interactions have been shown as a key regulatory mechanism for the precise genome regulation. Enhancers are only a small fraction of CREs and enhancers with poised state are not comprehensively studied. The lab uses the human beta globin cluster as a model cluster in which classic super-enhancers (LCR), repressors, and CREs with unknown function fine-tunes gene expression (Fig2). Importantly, the regulatory landscape of beta globin cluster extends out of canonical border of beta globin cluster between HS5 and 3’HS1 with our published study (Fig2). The lab is currently using omics tools like ChIP-Seq, CUT&RUN, in situ Hi-C, and micro-C, and regional capture micro-C (RCMC) to dissect the 3D genome regulation by CREs of this locus. We are currently utilizing the genome and 3D genome engineering approach to discover promising better and safer gene therapy for hemoglobinopathy.
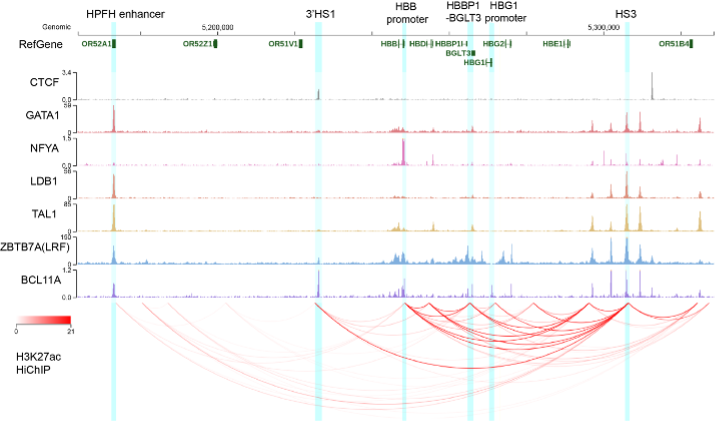
Fig 2. The CRE hub around human beta hemoglobin cluster in HUDEP2 cells
- Nuclear structure alterations in the leukemia and normal hematopoiesis
Nuclear morphology undergoes significant alterations during differentiation of multiple hematopoietic lineages (Fig.3). Yet, little is known on the nuclear structure alterations and gene expression during the hematopoietic lineage specification. We are using the advanced genomic technology like HiC, Micro-C to profile the 3D genomic interactions and during hematopoietic lineage specification. We previously identified the extreme long-range Polycomb loci interactions in the hematopoietic stem cells (Fig3.). We are currently profiling the extreme long-range Polycomb loci interactions and in function of these extreme long Polycomb interactions in cancer, particularly AML. We are also developing and adapting new long-read nanopore sequencing based genome foot printing methods, to profile genomic location of nuclear structural protein like nuclear lamina and nuclear pore complex during blood lineage specification.
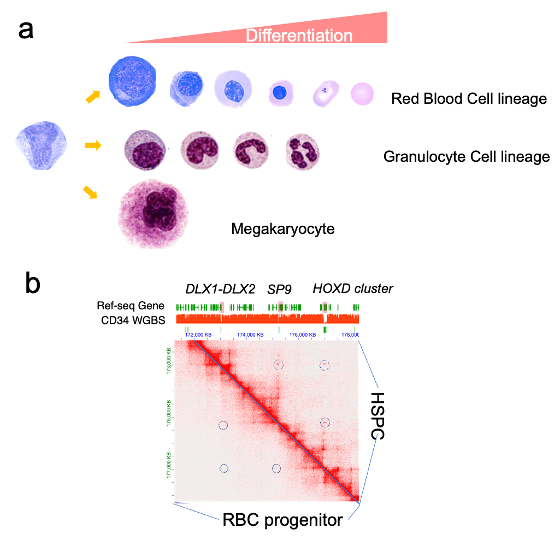
Fig 3. a. The morphology of cells during blood lineage specification (Modified from Yale Pathology, https://medcell.org/systems_cell_biology/haematopoiesis_lab.php). b. The HiC contact map of DNA methylation grand canyons in stem cells and RBC progenitors.
Selected publication:
- Wang XQ, Fan D, Liu Y, Han Q, Miao H, Wang X, Li Q, Chen D, Gore H, Himadewi P, Pfeifer GP, Grembecka J, Cierpicki T, Su J, Chong S, Wan L, Zhang X#. (# corresponding author). Mutant NPM1 hijacks transcriptional hubs to maintain pathogenic gene programs in acute myeloid leukemia. Cancer Discovery. (2023) 13 (3): 724–745. doi: 10.1158/2159-8290.CD-22-0424.
- Himadewi P, Wang XQ, Feng F, Gore H, Liu Y, Yu L, Kurita R, Nakamura Y, Pfeifer GP, Liu J, Zhang X# (# corresponding author). 3’HS1 CTCF binding site in human β-globin locus regulates fetal hemoglobin expression. Elife 2021;10:e
- Zhang X#, Jeong M, Huang X, Wang XQ, Wang X, Zhou W, Shamin M, Gore H, Himadewi P, Liu Y, Bochkov I, Reyes J, Doty M, Huang Y, Jung H, Heikamp E, Aiden A, Li W, Su J, Liberman-Aiden E#, Goodell MA#(#corresponding authors): Large DNA methylation nadirs anchor chromatin loops maintaining hematopoietic stem cell identity. Molecular Cell. Cover article. doi: 10.1016/j.molcel.2020.04.018.
Education and Training
BS
Biological Sciences, Fudan University
PhD
Molecular Human Genetics, Baylor College of Medicine
VARI Research Postdoctoral Fellow
Van Andel Research Institute
Graduate Program Affiliations
Genetics and Epigenetics
Cancer Biology
