
John Putkey, PhD
- Professor
- Vice-Chair of Administration
Areas of Interest
Research Interests
Calcium signaling, Calmodulin, PEP-19, NMR, Neurogranin
Molecular mechanisms of calcium signaling
Calcium is a key regulator of a wide spectrum of cellular processes. Numerous signals from hormones to action potentials induce fluctuations in intracellular calcium levels via the coordinated activities of numerous calcium channels and pumps. But calcium itself has no intrinsic activity, fluctuating calcium levels must be interpreted by regulatory proteins that bind calcium and then modulate down-stream processes via cascades of protein-protein interactions.
The most ubiquitous calcium regulatory protein is called calmodulin. It is relatively small with 4 EF-hand calcium binding sites. The primary sequence of calmodulin is highly conserved and it is essential for the viability of all eukaryotic cells from slime mold to humans due to its critical role in regulating cellular processes ranging from muscle contraction to neural transmission and cell division. Our lab has made hallmark contributions to the study of the biochemistry and biophysics of calmodulin. We were particularly interested in mechanisms that could control calmodulin activity other than binding calcium, and tune calmodulin to respond calcium signals of diverse amplitudes and frequencies. We discovered that the signaling properties of calmodulin are not dictated solely by calcium ions. A family of small intrinsically disordered proteins that include PEP-19 and neurogranin bind to calmodulin and modulate how it senses calcium signals. Using a combination of biophysical techniques and NMR, we showed that PEP-19 modifies the calcium binding properties of calmodulin by electrostatically steering calcium to binding sites on calmodulin (see Figure), while neurogranin tunes the rate of association of calmodulin with its target proteins. Modulation of calmodulin activity by these small, disordered regulators of calmodulin signaling has broad implications on a wide spectrum of normal and pathological conditions, including cancer.
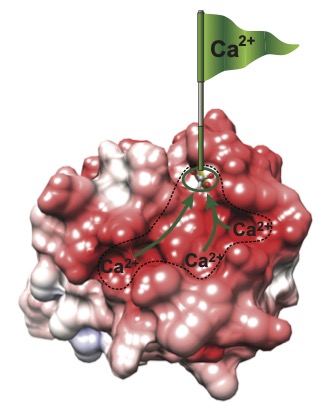
Negative electrostatic surface potential (red) contributed by PEP-19 steers calcium to binding sites in calmodulin
Selected Publications
Kleerekoper, Q.K., and Putkey, J.A., (2009) PEP-19 is an intrinsically disordered regulator of calmodulin signaling. J. Biol. Chem. 284: 7455-7464. PMCID: PMC2658041
Wang, W. and Putkey, JA (2016) PEP-19 modulates calcium binding to calmodulin by electrostatic steering. Nature Communications. 7:13583. doi: 10.1038/ncomms13583.
Wang, X, Gorfe, A.A., and Putkey, J.A. (2021) Antipsychotic phenothiazine drugs bind to KRAS in vitro. J. Biomol. NMR 75: 233-244.
Putkey, J, Hoffman, L, Berka, V, and Wang X (2024) Neurogranin modulates the rate of association between calmodulin and target peptides. Biophys. J. 123:1676-1686. 1016/j.bpj.2024.05.010
Education and Training
Undergraduate
Biochemistry, University of California, Berkeley
PhD
Biochemistry, University of California, Riverside
Damon Runyon-Walter Winchell Postdoctoral Fellow
Cell Biology, Baylor college of Medicine
Graduate Program Affiliations
Molecular and Translational Biology