
Wenbo Li, PhD
- Associate Professor
- CPRIT Scholar, Cancer Prevention Research Institute of Texas
Areas of Interest
Research Interests
Genomics, epigenomics, bioinformatics, long noncoding RNAs, enhancer RNAs (eRNAs), enhancers, three-dimensional genome architecture, cohesin and condensin, genomic variations/mutations, RNA binding proteins.
New frontiers of gene regulation: charting the function and regulation of noncoding RNA/DNA in the human genome
A decade after the completion of the Human Genome Project, a new challenge now is to decipher the function and regulation, as well as the complex interplay, of millions of regulatory elements in the human genome. These elements together govern almost all basic cellular functions ranging from DNA replication and genome integrity to gene transcriptional control. Importantly, the mutations of noncoding regulatory elements, such as enhancers and promoters, are associated with various diseases, and constitute the majority of genomic variations/mutations in humans as compared to much-studied mutations of protein coding genes. However, our knowledge of the molecular mechanisms underlying genomic regulatory elements remains incomplete, which prevents the solving of this new challenge.
Dr. Li’s laboratory attempts to contribute to solve this challenge by focusing on three major directions (see below). Highly inter-disciplinary approaches will be utilized which include those of biochemistry and molecular biology, epigenetics/epigenomics and bioinformatics.
Elucidating the molecular functions of enhancer-derived noncoding RNAs (eRNAs)
Networks of regulatory enhancers dictate distinct cell identities and cellular responses to diverse signals by instructing precise spatiotemporal patterns of gene expression. However, 35 years after their discovery, enhancer functions and mechanisms remain incompletely understood. Intriguingly, recent evidence suggests that many, if not all, functional enhancers are themselves transcription units, generating eRNAs. This observation provides a fundamental insight into the inter-regulation between enhancers and promoters, which can both act as transcription units; it also raises crucial questions regarding the regulation of the enhancer transcription process and potential biological roles of non-coding eRNAs (Fig.1).
We will use biochemical methods such as antisense oligo based RNA pull down coupled with mass spectrometry, and omics approaches such as GRO-seq (global run-on sequencing) and CLIP-seq (cross-linking immunoprecipitation and sequencing), as well as cutting-edge RNA-DNA interaction and sequencing assays to study the targets of selective eRNAs, their protein partners, potential chemical RNA modifications and functional roles in gene regulation, or in the organization of specific genome architectures and in human disease such as cancer.
Characterizing the three-dimensional genome architecture in gene regulation and cancer
An amazing feature of eukaryotic nuclei is that as long as 2 meters of DNA in linear length (3 billion base pairs in humans) needs to be packaged into a space of les than 10 μm a diameter, while the information stored in all regions of this stretch of DNA can still be very quickly (in minutes or seconds) and effectively retrieved during the daily activity of a cell or an organism. (This process is so effective and precise that it almost for sure outperforms The Google search engine!). To characterize the three-dimensional (3D) genome architecture will offer new insights into the process as to how regulatory elements talk to each other in this highly folded space. Importantly, it is increasingly realized that many human diseases are associated with the disruption of proper chromatin architecture or nuclear locations of genomic regulatory elements, rasing the possibility that by understanding and modulating the genome architecture, there could be innovative ways to interfere with human disease. For this specific direction, I have previously worked extensively on important chromatin architecture regulators such as cohesin and condensin complexes in hormonal gene regulation. Intriguingly, these complexes were recently found highly mutated in many human cancers, without clear mechanisms being yet deciphered. We will use system-wide genome architecture assays named 4C-seq and HiChIP/PLAC-seq, extensive bioinformatic analyses and mathematic modeling to understand how these molecules intervene the 3D genome, and how their mutations lead to cancer.
Understanding the noncoding genome variations/mutations in human cancer
The study of the previous two directions aim to elucidate the basic mechanisms underlying the RNA or DNA elements encoded in the human genome. However, a major importance of mechanistic studies is that they will ultimately lead to innovative ways to intervene human disease. With the advent of cheaper and more efficient genome sequencing technologies, genome modifying tools (e.g. Crispr/Cas9 or C2c2) and more comprehensive knowledge of human genome mutations and variations (e.g. GWAS and TCGA databases), we hope to link the mechanisms of eRNA and 3D genome with poorly understood human mutations in the noncoding regions. For examples, the mutations of enhancers may be linked to gain or loss of enhancer activity, or alteration of enhancer-promoter looping events. We also wish to mine datasets of genomes of different species during evolution to gain insights into the functional drifting of regulatory elements such as enhancers. We envision that these directions will lead to unexpected findings and novel perspectives to therapeutically target human disease such as cancer.
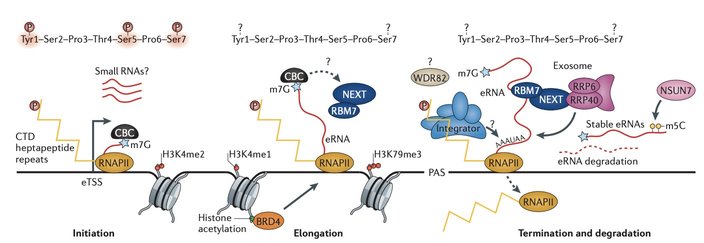
The transcription process of enhancers and some recently uncovered regulators.
Selected Publications
Li W, Hu Y, Oh S, Ma Q, Merkurjev D, Song X, Zhou X, Liu Z, Tanasa B, He X, Chen AY, Ohgi K, Zhang J, Liu W, Rosenfeld MG. 2015. Condensin I and II Complexes License Full Estrogen Receptor α-Dependent Enhancer Activation. Mol Cell. 59: 188-202.
Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, Oh S, Kim HS, Glass CK, Rosenfeld MG. 2013. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 498:516-20.
Li W, Notani D, Rosenfeld MG. 2016. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet.17:207-23.
Education and Training
BS
Zhejiang University
PhD
National University of Singapore
Postdoctoral Fellow
University of California San Diego
Graduate Program Affiliations
Genetics and Epigenetics
Molecular and Translational Biology