Research
Lipid-Dependent Membrane Protein Topological Organization
Membrane protein topology and assembly are governed by structural principles and topological rules and directed by topogenic signals and sequences in the nascent polypeptide chain that are recognized and decoded not only by the translocon but also by the given lipid profile. The dynamic Charge Balance Rule as an extension of the static Positive Inside Rule was proposed by us to provide a mechanistic understainding of membrane protein structura; dynamics and generation of membrane protein structural duality and heterogeneity.
The goal of the first project in the lab is to investigate the molecular mechanism and physiological significance of lipid-dependent membrane protein topogenesis. We are testing the novel Charge Balance Rule for different proteins and lipid profiles in vivo and in vitro in order to establish the physiological significance of conformational and topological heterogeneity, co-translational, post-translational and post-insertional dynamic changes in topological organization of membrane proteins. We employed bacterial cells, mammalian cells and their organelles, as well as cancer cells where changes in lipid asymmetry and external environment occur regularly. In addition, we test the Charge Balance Rule in vivo and in silico to verify the concept of the funnel-like intermolecular energy landscape of lipid–protein transient interactions by using of a series of in vivo “translocation force” measurements and computational experiments
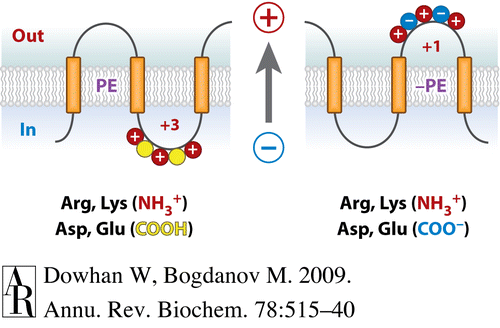
Fig. 1. Charge Balance Rule for membrane protein assembly.
Dynamic lipid asymmetry
Why is the cell membrane asymmetric? When proteins and lipids are synthesized in the cell, they are inserted into the membrane in an asymmetric fashion. Although this arrangement becomes a main architectural principle for most if not all living biological membranes, we are still far from full understanding of the physiological significance and detailed molecular mechanisms by which membrane phospholipid asymmetry is generated, maintained and modulated. What is the reason for the cell membrane to be asymmetric in biogenic- and non-biogenic cells, healthy and cancer cells? Although biological membranes are almost universally asymmetric, the asymmetry is not absolute and symmetric biological membranes appear to exist in nature. Therefore, new explanatory hypotheses and experimental models are needed to understand how widely transmembrane heterogeneity is distributed among different forms of life. We pioneered methods to investigate individual phospholipid topography (transmembrane sidedness) using novel assays to establish the function of individual lipid enzymes and lipids in such complex processes as translocation of lipids and proteins across the lipid bilayer, membrane protein folding and topogenesis, generation and maintenance of membrane asymmetry a well as adaptation of bacterial cells to different environments. Utilizing two vectorial probes with different membrane penetrating and chemical properties, we report novel methodology to determine the transmembrane distribution of PE and other amino lipids (tentatively designated as vectorial lipidomics). Characterization of covalently labeled lipids with independent radiolabeled, normal-phase LC/MS/MS, TLC elution–based, and TLC-less spectrophotometric assays allowed us to determine asymmetrical, dynamic, and cell shape–dependent distribution of the aminophospholipid head groups and acyl chaines. In addtion, we assessed physical leaflet-specific asymmetry by lipid-order and leaflet specific advanced fluorescent probes in the biogenic inner membrane (IM) of diderm Gram-negative bacteria.
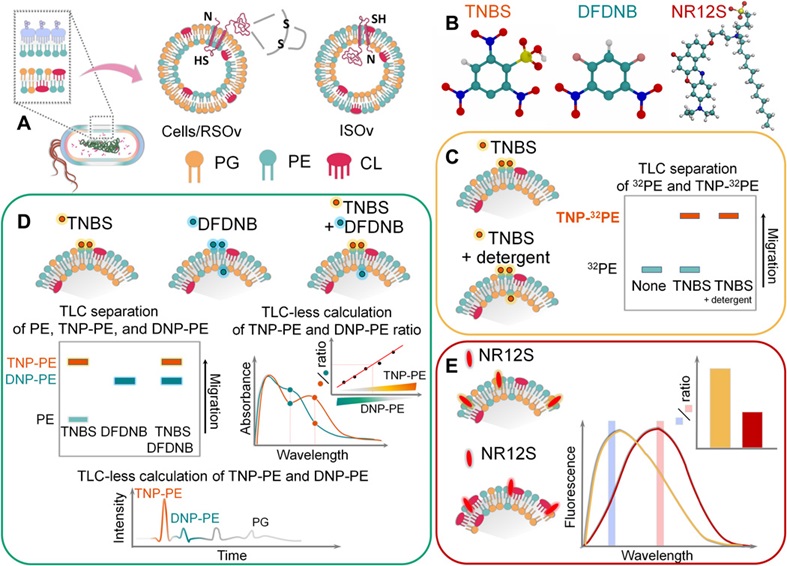
Fig. 2. Vectorial lipidomics. Characterization of covalently labeled lipids with independent radiolabeled, normal-phase LC/MS/MS, TLC elution–based, and TLC- less spectrophotometric assays. Bogdanov et al., Sci Adv.6(23):eaaz6333, 2020.
Still much remains to be discovered and several controversies remain to be resolved. If the asymmetric IM of bacteria is dispensable for cell viability, it may be difficult or even impossible to genetically identify flippase candidates by searching for synthetic lethal interactions and loss of function mutations unless new cellular circumstances in stressed cells will be uncovered. However, the biochemical reconstitution and search for in vivoconditions which can modulate an IM asymmetry represent a very reasonable starting point. Although PE is asymmetrically distributed across the IM, currently it is not known whether PG or CL are asymmetrically distributed across the IM and whether their sidedness is physiologically important. How are the compositional (head and acyl groups), molecular (combination of acyls with different length and unsaturation within the same lipid class of molecules) and physical (lipid order and packing) lipid asymmetries regulated in biogenic cytoplasmic membranes? An identification of the mechanism (flippase-free or flippase-guided) controlling the asymmetric distribution of lipids across the bacterial IM is our guiding factor for the future research focused on diderm bacteria.
A dynamic view of lipid and protein organization reveals previously unrecognized possibilities for cellular regulation and understanding of disease states resulting from mis-folded and mis-oriented proteins. The understanding of the principles governing lipid-dependent assembly and organization of membrane proteins could be useful in developing of novel therapeutic approach for protein disorders where the conformation or topology of a membrane protein needs to be modified either directly or indirectly through membrane-lipid engineering.
Selected publications
Lipid-assisted Membrane Protein Folding: Lipids as Molecular Chaperones (Lipochaperones).
- Bogdanov M, Sun J, Kaback HR and Dowhan W: A phospholipid acts as a chaperone in assembly of membrane transport protein. Biol. Chem.271: 11615-11618, 1996.
- Bogdanov M and Dowhan W: Lipid-assisted protein folding J. Biol. Chem. 274: 36827-36830, 1999.
- Bogdanov M, Heacock P, Guan Z and Dowhan W: Plasticity of lipid-protein interactions in the function and topogenesis of the membrane protein lactose permease from Escherichia coli. Natl. Acad. Sci. USA.107(34):15057-62, 2010.
Lipid-Dependent Membrane Protein Topological Organization
- Bogdanov M, Xie J, Heacock P and Dowhan, W: To flip or not to flip: protein–lipid charge interactions are a determinant of final membrane protein topology J. Cell Biology. 182: 925-935, 2008.
- Dowhan W and Bogdanov M: Lipid-dependent topogenesis. Rev. Biochem. 78: 515-540, 2009.
- Bogdanov M, Dowhan W, Vitrac H: Lipids and topological rules governing membrane protein assembly. Biochim Biophys Acta. 1843(8): 1475-1488, 2014..
- Bogdanov, M., Vitrac. H., Dowhan, W. Flip-flopping Membrane Proteins: How the Charge Balance Rule Governs Dynamic Membrane Protein Topology. Biogenesis of Fatty Acids, Lipids and Membranes (Geiger, O., Ed) Second Edition, Elsevier Press, Amsterdam, 2018.
- Dowhan, W., Vitrac, H. and Bogdanov, M: Lipid assisted membrane protein folding and topogenesis (Special issue honoring Günther Blobel). Protein J. 38: 274-88, 2019
- Bogdanov M. Exploring uniform, dual and dynamic topologies of membrane proteins by substituted cysteine accessibility method (SCAMTM) Methods in Molecular Biology. 2715:121-157, 2023
Dynamic lipid asymmetry
- Bogdanov, M., Pyrshev, K., Yesylevskyy, S., Ryabichko, S., Boiko, V., Ivanchenko, P., Kiyamova, R, Guan, Z., Ramseyer , C., Dowhan, W. Phospholipid distribution in the cytoplasmic membrane of Gram-negative bacteria is highly asymmetric, dynamic, and cell shape-dependent Sci Adv. 6(23):eaaz6333, 2020
- Bogdanov M. The power and challenge of lipid (a)symmetry across the membrane and cell. Emerg Top Life Sci. 7(1):1-6, 2023
- Bogdanov M. Renovating a double fence with or without notifying the next door and across the street neighbors: why the biogenic cytoplasmic membrane of Gram-negative bacteria display asymmetry? Emerg Top Life Sci. 7(1):137-150, 2023
Role of Gram-negative bacterial envelope remodeling in the development of resistance to antibiotics, intracellular survival and ability to resist innate immunity of the host
- Zheng, L., Lin, Y., Lu, S, Zhang, J, Bogdanov M. Biogenesis, transport and remodeling of lysophospholipids in Gram-negative bacteria. Biochim Biophys Acta. S1388-1981(16) 30332-8, 2016.
- Sanina, N., Pomazenkova, L., Bakholdina, S., Chopenko, N., Zabolotnaya, A., Reutov, V., Stenkova, A., Bystritskaya, E., and Bogdanov, M. Relationship between adaptive changing of lysophosphatidylehanolamine content in bacterial envelope and ampicillin sensitivity of Yersinia pseudotuberculosis. Journal of Molecular Microbiology and Biotechnology, 28 (5) 236-239, 2019.