
Irina I. Serysheva, PhD
- Professor
- Jesse H. Jones Chair
- Director, Structural Biology Imaging Center
Areas of Interest
Research Interests
Structure-Function of Ion Channels
Cellular Ca2+ Signaling
Membrane Transport
Electron Cryo-microscopy
Image Processing and 3D Reconstruction
Structure Determination of Macromolecular Complexes
Structural Proteomics
Structure and function of integral membrane proteins
Our research aims to understand molecular mechanisms underlying transport of molecules into and out of the cell across the surface membrane, or between different intracellular compartments through structure-functional studies of integral membrane proteins known as ion channels, and the macromolecular complexes they form. Ion channels regulate many diverse biological functions that include muscle contraction, hormone secretion, gene transcription, metabolic regulation, neurotransmitter release, fertilization and apoptosis. The knowledge about the three dimensional (3D) architecture of ion channels is required to understand molecular basis of ion channel gating (opening/closing process), and how this process is controlled by a wide variety of endogenous molecules and pharmacological modifies. To answer these questions we use a combination of electron microscopy and computer reconstruction techniques in conjunction with biochemical, electrophysiological and molecular biological approaches. Our structure research efforts include:
- purification of ion channels from natural sources or from high-level expression systems;
- electron cryomicroscopy (cryo-EM) of the purified channel assemblies;
- computer image processing and 3D reconstruction;
- structure analysis and annotation using combination of visualization and computational tools;
- prediction of functional roles of the identified structural domains via bioinformatics.
Recent focus has been on structural analysis of Ca2+ channels that mediate ligand-gated release of Ca2+ from intracellular stores: the ryanodine-sensitive Ca2+ release channel (RyR), the primary Ca2+ release channel in muscle cells, and the inositol 1,4,5-trisphosphate-sensitive Ca2+ release channel (IP3R), localized in the endoplasmic reticulum. Both channels are large tetrameric protein complexes with a molecular mass of ~2.3 MDa for RyRs and 1.2 MDa for IP3Rs. Defects in these channel proteins cause abnormal regulation of cell Ca2+ level underlying numerous human diseases: Malignant Hyperthermia, Central Core disease, cardiac hypertrophy, heart failure, hereditary ataxias, Huntington’s disease, Alzheimer’s disease, osteoporosis, atherosclerosis and some migraines.
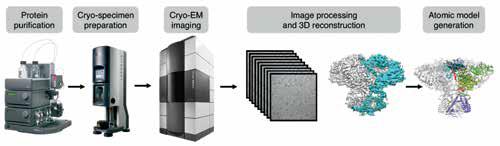
The cryo-EM structure determination pipeline that allows for solving atomic resolution structures of biological assemblies in different functional states.
Selected Publications
Gating machinery of InsP3R channels revealed by electron cryomicroscopy. Fan G, Baker ML, Wang Z, Baker MR, Sinyagovskiy PA, Chiu W, Ludtke SJ, Serysheva II. Nature. 2015 Oct 12.
Cryo-EM reveals ligand induced allostery underlying InsP3R channel gating. Fan G, Baker MR, Wang Z, Seryshev AB, Ludtke SJ, Baker ML, Serysheva II. Cell Res. 2018 Dec;28(12):1158-1170.
Cryo-EM structure of type 1 IP3R channel in a lipid bilayer. Baker, M.R., Fan, G., Seryshev, A.B., Agosto M.A., Baker M.L., Serysheva I.I. Commun Biol 4, 625 (2021).
Conformational motions and ligand-binding underlying gating and regulation of IP3R channel. Fan G, Baker MR, Terry LE, Arige V, Chen M, Seryshev AB, Baker ML, Ludtke SJ, Yule DI, Serysheva II. Nat Commun. 2022 Nov 14;13(1):6942.
Understanding IP3R channels: From structural underpinnings to ligand-dependent conformational landscape. Baker MR, Fan G, Arige V, Yule DI, Serysheva II. Cell Calcium. 2023 Jun 22;114:102770.
Education and Training
BS
Biochemistry, MV Lomonosov State University
MS
Virology, MV Lomonosov State University
PhD
Biochemistry, AN Bach Institute of Biochemistry
Postdoctoral Fellow
Molecular Biology and Biophysics, AN Bach Institute of Biochemistry
Graduate Program Affiliations
Molecular and Translational Biology