SAM (Significance Analysis of Microarray) Results
Description of the problem:
Number of classes: 2
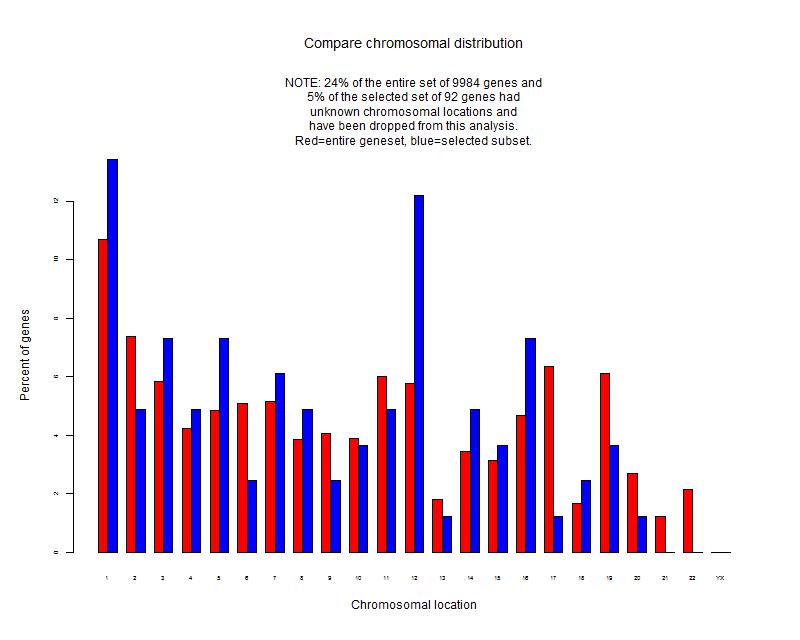
Number of genes that passed filtering criteria: 9984
Type of test used: SAM – Significance analysis of Microarray
Column of the Experiment Descriptors sheet that defines class variable : AFP_100
Target proportion of false discoveries : 0.1
Number of permutations : 100
Percentile for determining called genes that are false : 80
Genes which discriminate among classes:
Table 1
Class 1: 0; Class 2: 1.
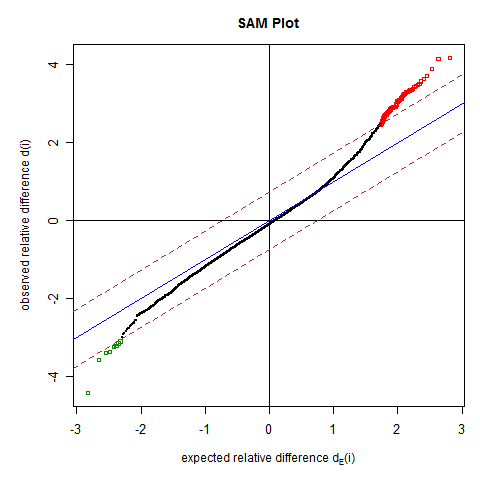
There are 92 genes significant by SAM
The estimated false discovery rate among the 92 significant genes is 0.08236 .
The delta value used to identify the significant genes is 0.73681 .
The fudge factor for standard deviation is computed as 0.01517 .
d(i) is the observed relative difference for two-class problem. Click the HELP link at the top of the page for more information.
| d(i) | Geom mean of ratios in class 1 | Geom mean of ratios in class 2 | Fold-change | Unique ID | Annotations | Clone ID | Accession | Symbol | Entrez ID | Defined Gene list | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 4.177 | 0.92 | 0.64 | 1.45 | 161146 | 161146 Info | Incyte PD:2799664 | U89281 | HSD17B6 | 8630 | |
| 2 | 4.144 | 0.95 | 0.69 | 1.36 | 168187 | 168187 Info | Incyte PD:2099420 | AI310515 | ACSM2B | 348158 | |
| 3 | 3.892 | 0.85 | 0.59 | 1.43 | 161801 | 161801 Info | Incyte PD:5034123 | L21893 | SLC10A1 | 6554 | |
| 4 | 3.719 | 0.78 | 0.56 | 1.38 | 166007 | 166007 Info | Incyte PD:606122 | NM_000353 | TAT | 6898 | Alkaloid biosynthesis I, Novobiocin biosynthesis, Phenylalanine metabolism, Phenylalanine, tyrosine and tryptophan biosynthesis, Tyrosine metabolism, immunology |
| 5 | 3.64 | 0.94 | 0.66 | 1.42 | 167207 | 167207 Info | Incyte PD:2590673 | U39967 | FMO3 | 2328 | immunology |
| 6 | 3.573 | 0.91 | 0.71 | 1.28 | 168452 | 168452 Info | Incyte PD:1297179 | NM_005956 | MTHFD1 | 4522 | Glyoxylate and dicarboxylate metabolism, One carbon pool by folate |
| 7 | 3.515 | 0.64 | 0.47 | 1.37 | 169374 | 169374 Info | Incyte PD:2513602 | NM_002150 | HPD | 3242 | Phenylalanine metabolism, Tyrosine metabolism |
| 8 | 3.485 | 0.78 | 0.62 | 1.26 | 168756 | 168756 Info | Incyte PD:1794046 | M93405 | ALDH6A1 | 4329 | Inositol metabolism, Propanoate metabolism, Valine, leucine and isoleucine degradation |
| 9 | 3.459 | 1.08 | 0.86 | 1.25 | 169717 | 169717 Info | Incyte PD:1846226 | AA970835 | PGRMC1 | 10857 | |
| 10 | 3.431 | 0.93 | 0.65 | 1.43 | 167792 | 167792 Info | Incyte PD:1308102 | D00230 | OTC | 5009 | Arginine and proline metabolism, Urea cycle and metabolism of amino groups, immunology, metabolism |
| 11 | 3.385 | 0.8 | 0.56 | 1.42 | 164998 | 164998 Info | Incyte PD:1633340 | Y15793 | CPS1 | 1373 | Arginine and proline metabolism, Glutamate metabolism, Nitrogen metabolism, Urea cycle and metabolism of amino groups, immunology |
| 12 | 3.351 | 0.86 | 0.71 | 1.22 | 165794 | 165794 Info | Incyte PD:234123 | AI301066 | EPHX2 | 2053 | Eicosanoid Metabolism, Arachidonic acid metabolism, Tetrachloroethene degradation, immunology, metabolism, pharmacology |
| 13 | 3.348 | 1.13 | 0.92 | 1.23 | 160308 | 160308 Info | Incyte PD:1445238 | NM_014333 | CADM1 | 23705 | Cell adhesion molecules (CAMs) |
| 14 | 3.346 | 0.83 | 0.57 | 1.47 | 161478 | 161478 Info | Incyte PD:3752047 | AB008775 | AQP9 | 366 | |
| 15 | 3.323 | 0.95 | 0.76 | 1.25 | 165324 | 165324 Info | Incyte PD:1833801 | BE393339 | PXMP2 | 5827 | |
| 16 | 3.307 | 0.99 | 0.83 | 1.2 | 168366 | 168366 Info | Incyte PD:1287840 | AI675901 | SCP2 | 6342 | PPAR signaling pathway |
| 17 | 3.303 | 0.81 | 0.59 | 1.38 | 166095 | 166095 Info | Incyte PD:1311471 | AA113231 | CPS1 | 1373 | Arginine and proline metabolism, Glutamate metabolism, Nitrogen metabolism, Urea cycle and metabolism of amino groups, immunology |
| 18 | 3.291 | 0.73 | 0.57 | 1.28 | 165233 | 165233 Info | Incyte PD:1427681 | H94887 | GLYAT | 10249 | |
| 19 | 3.258 | 1.01 | 0.86 | 1.17 | 165777 | 165777 Info | Incyte PD:685416 | M36429 | GNB2 | 2783 | |
| 20 | 3.252 | 1 | 0.89 | 1.13 | 161491 | 161491 Info | Incyte PD:2554470 | X83441 | LIG4 | 3981 | DNA_adducts, DNA_damage |
| 21 | 3.222 | 0.9 | 0.7 | 1.28 | 166778 | 166778 Info | Incyte PD:1631511 | X92720 | PCK2 | 5106 | Adipocytokine signaling pathway, Citrate cycle (TCA cycle), Insulin signaling pathway, PPAR signaling pathway, Pyruvate metabolism |
| 22 | 3.198 | 0.95 | 0.74 | 1.29 | 162315 | 162315 Info | Incyte PD:489032 | D43704 | CACNB3 | 784 | MAPK signaling pathway |
| 23 | 3.168 | 0.96 | 0.81 | 1.18 | 166910 | 166910 Info | Incyte PD:2058620 | AI309555 | ADI1 | 55256 | |
| 24 | 3.138 | 0.93 | 0.77 | 1.22 | 167281 | 167281 Info | Incyte PD:546754 | NM_003718 | CDK13 | 8621 | |
| 25 | 3.137 | 0.91 | 0.77 | 1.18 | 164136 | 164136 Info | Incyte PD:1300469 | M74096 | ACADL | 33 | beta-Alanine metabolism, Fatty acid metabolism, PPAR signaling pathway, Propanoate metabolism, Valine, leucine and isoleucine degradation, immunology |
| 26 | 3.11 | 0.88 | 0.69 | 1.27 | 160425 | 160425 Info | Incyte PD:666190 | S52784 | CTH | 1491 | Cysteine metabolism, Glycine, serine and threonine metabolism, Methionine metabolism, Nitrogen metabolism, Selenoamino acid metabolism, misc |
| 27 | 3.108 | 0.91 | 0.68 | 1.33 | 165825 | 165825 Info | Incyte PD:1902152 | AW627651 | SDS | 10993 | Cysteine metabolism, Glycine, serine and threonine metabolism |
| 28 | 3.092 | 0.74 | 0.5 | 1.49 | 161296 | 161296 Info | Incyte PD:1669534 | K01763 | HP | 3240 | immunology, misc |
| 29 | 3.088 | 0.95 | 0.82 | 1.15 | 161511 | 161511 Info | Incyte PD:3271263 | X12910 | ATP1A3 | 478 | |
| 30 | 3.084 | 0.82 | 0.66 | 1.25 | 166057 | 166057 Info | Incyte PD:400429 | NM_000306 | POU1F1 | 5449 | transcription |
| 31 | 3.064 | 0.94 | 0.72 | 1.31 | 164992 | 164992 Info | Incyte PD:4244645 | AI791352 | DAO | 1610 | Arginine and proline metabolism, D-Arginine and D-ornithine metabolism, Glycine, serine and threonine metabolism, immunology |
| 32 | 3.06 | 0.86 | 0.66 | 1.29 | 169679 | 169679 Info | Incyte PD:87242 | ||||
| 33 | 2.989 | 0.92 | 0.81 | 1.14 | 163485 | 163485 Info | Incyte PD:3561805 | NM_001942 | DSG1 | 1828 | Cell Communication, immunology |
| 34 | 2.984 | 0.89 | 0.72 | 1.23 | 160233 | 160233 Info | Incyte PD:614679 | Y12735 | DYRK3 | 8444 | signal_transduction |
| 35 | 2.975 | 0.99 | 0.86 | 1.15 | 166974 | 166974 Info | Incyte PD:1702350 | AW025439 | GADD45A | 1647 | ATM Signaling Pathway, Cell Cycle: G2/M Checkpoint, Hypoxia and p53 in the Cardiovascular system, p53 Signaling Pathway, Cell cycle, MAPK signaling pathway |
| 36 | 2.925 | 1 | 0.82 | 1.21 | 167196 | 167196 Info | Incyte PD:1922001 | AW246790 | BDH1 | 622 | Butanoate metabolism, Synthesis and degradation of ketone bodies |
| 37 | 2.922 | 1 | 0.9 | 1.12 | 163580 | 163580 Info | Incyte PD:3200341 | M15169 | ADRB2 | 154 | Corticosteroids and cardioprotection, Cystic fibrosis transmembrane conductance regulator (CFTR) and beta 2 adrenergic receptor (b2AR) pathway, Phospholipase C-epsilon pathway, Calcium signaling pathway, Neuroactive ligand-receptor interaction, behavior, immunology |
| 38 | 2.918 | 0.92 | 0.75 | 1.22 | 160471 | 160471 Info | Incyte PD:532254 | X95384 | HRSP12 | 10247 | |
| 39 | 2.916 | 0.87 | 0.68 | 1.28 | 169589 | 169589 Info | Incyte PD:911015 | U63008 | HGD | 3081 | Styrene degradation, Tyrosine metabolism |
| 40 | 2.913 | 0.92 | 0.75 | 1.23 | 159780 | 159780 Info | Incyte PD:1601609 | AJ001319 | MPDZ | 8777 | Tight junction |
| 41 | 2.912 | 0.7 | 0.51 | 1.37 | 161003 | 161003 Info | Incyte PD:2373085 | M24317 | ADH1B | 125 | 1- and 2-Methylnaphthalene degradation, Bile acid biosynthesis, Fatty acid metabolism, Glycerolipid metabolism, Glycolysis / Gluconeogenesis, Metabolism of xenobiotics by cytochrome P450, Tyrosine metabolism |
| 42 | 2.909 | 0.89 | 0.73 | 1.23 | 164412 | 164412 Info | Incyte PD:1804662 | NM_003869 | CES2 | 8824 | |
| 43 | 2.908 | 0.7 | 0.52 | 1.34 | 161259 | 161259 Info | Incyte PD:2516815 | X98332 | SLC22A1 | 6580 | |
| 44 | 2.896 | 0.74 | 0.55 | 1.34 | 160174 | 160174 Info | Incyte PD:166909 | J05064 | C6 | 729 | Alternative Complement Pathway, Cells and Molecules involved in local acute inflammatory response, Classical Complement Pathway, Complement Pathway, Lectin Induced Complement Pathway, Complement and coagulation cascades, immunology |
| 45 | 2.89 | 0.99 | 0.89 | 1.11 | 161323 | 161323 Info | Incyte PD:3419068 | AF071542 | SEMA7A | 8482 | Axon guidance |
| 46 | 2.887 | 0.91 | 0.77 | 1.18 | 163533 | 163533 Info | Incyte PD:1930954 | AA805411 | ECHDC2 | 55268 | |
| 47 | 2.886 | 0.93 | 0.81 | 1.15 | 162878 | 162878 Info | Incyte PD:1805089 | AI192537 | TMEM56 | 148534 | |
| 48 | 2.858 | 0.77 | 0.65 | 1.17 | 163887 | 163887 Info | Incyte PD:1595081 | M59979 | PTGS1 | 5742 | Aspirin Blocks Signaling Pathway Involved in Platelet Activation, Eicosanoid Metabolism, Mechanism of Acetaminophen Activity and Toxicity, Arachidonic acid metabolism, immunology |
| 49 | 2.857 | 0.75 | 0.6 | 1.24 | 159969 | 159969 Info | Incyte PD:1664320 | M14058 | C1R | 715 | Classical Complement Pathway, Complement Pathway, Complement and coagulation cascades, immunology |
| 50 | 2.838 | 0.91 | 0.74 | 1.23 | 161484 | 161484 Info | Incyte PD:2515733 | AF054821 | CYP4F3 | 4051 | Arachidonic acid metabolism, pharmacology |
| 51 | 2.829 | 1.04 | 0.87 | 1.2 | 165274 | 165274 Info | Incyte PD:1740924 | AA203542 | SLC6A1 | 6529 | |
| 52 | 2.828 | 0.78 | 0.54 | 1.44 | 162599 | 162599 Info | Incyte PD:1740889 | M69197 | HPR | 3250///3240 | immunology |
| 53 | 2.813 | 1 | 0.88 | 1.13 | 164597 | 164597 Info | Incyte PD:1923117 | AI269221 | C8orf40 | 114926 | |
| 54 | 2.807 | 0.83 | 0.64 | 1.28 | 169027 | 169027 Info | Incyte PD:2516665 | U32576 | APOC4 | 346 | |
| 55 | 2.795 | 1.03 | 0.9 | 1.15 | 160100 | 160100 Info | Incyte PD:1811220 | NM_001182 | ALDH7A1 | 501 | Arginine and proline metabolism, Ascorbate and aldarate metabolism, beta-Alanine metabolism, Bile acid biosynthesis, Butanoate metabolism, Fatty acid metabolism, Glycerolipid metabolism, Glycolysis / Gluconeogenesis, Histidine metabolism, Limonene and pinene degradation, Lysine degradation, Propanoate metabolism, Pyruvate metabolism, Tryptophan metabolism, Valine, leucine and isoleucine degradation |
| 56 | 2.794 | 0.97 | 0.84 | 1.16 | 164495 | 164495 Info | Incyte PD:1968776 | AW275347 | ACACB | 32 | Adipocytokine signaling pathway, Fatty acid biosynthesis, Insulin signaling pathway, Propanoate metabolism, Pyruvate metabolism |
| 57 | 2.793 | 0.84 | 0.7 | 1.2 | 165554 | 165554 Info | Incyte PD:943569 | AA707391 | ALAS1 | 211 | Hemoglobin\’s Chaperone, Glycine, serine and threonine metabolism |
| 58 | 2.792 | 0.79 | 0.59 | 1.33 | 164296 | 164296 Info | Incyte PD:2514507 | NM_000040 | APOC3 | 345 | PPAR signaling pathway, immunology |
| 59 | 2.758 | 1.05 | 0.91 | 1.15 | 166966 | 166966 Info | Incyte PD:1638850 | NM_004493 | HSD17B10 | 3028 | Butanoate metabolism, Caprolactam degradation, Fatty acid elongation in mitochondria, Fatty acid metabolism, Lysine degradation, Tryptophan metabolism, Valine, leucine and isoleucine degradation |
| 60 | 2.741 | 0.99 | 0.85 | 1.16 | 169282 | 169282 Info | Incyte PD:88935 | ||||
| 61 | 2.739 | 0.81 | 0.61 | 1.32 | 162415 | 162415 Info | Incyte PD:139449 | NM_001713 | BHMT | 635 | Glycine, serine and threonine metabolism, Methionine metabolism, misc |
| 62 | 2.732 | 0.97 | 0.87 | 1.12 | 168483 | 168483 Info | Incyte PD:1938951 | AI536745 | FAM134B | 54463 | |
| 63 | 2.731 | 0.97 | 0.85 | 1.15 | 167750 | 167750 Info | Incyte PD:2840195 | NM_001123 | ADK | 132 | Purine metabolism, cell_cycle, cell_signaling, immunology, signal_transduction |
| 64 | 2.722 | 0.97 | 0.85 | 1.13 | 165041 | 165041 Info | Incyte PD:259054 | AL034562 | LOC727993 | 727993///140885///5173 | |
| 65 | 2.713 | 0.77 | 0.64 | 1.21 | 160017 | 160017 Info | Incyte PD:1664796 | D85510 | MMP15 | 4324 | immunology |
| 66 | 2.711 | 0.94 | 0.82 | 1.14 | 161416 | 161416 Info | Incyte PD:1957615 | X52638 | PFKFB1 | 5207 | Fructose and mannose metabolism |
| 67 | 2.686 | 0.9 | 0.63 | 1.43 | 162311 | 162311 Info | Incyte PD:480059 | NM_000488 | SERPINC1 | 462 | Acute Myocardial Infarction, Extrinsic Prothrombin Activation Pathway, Intrinsic Prothrombin Activation Pathway, Complement and coagulation cascades |
| 68 | 2.686 | 0.68 | 0.55 | 1.24 | 164943 | 164943 Info | Incyte PD:168865 | M17398 | CYP2C8 | 1558 | Arachidonic acid metabolism, Linoleic acid metabolism, Metabolism of xenobiotics by cytochrome P450 |
| 69 | 2.682 | 0.79 | 0.64 | 1.24 | 161326 | 161326 Info | Incyte PD:3146420 | AF057034 | RDH16 | 8608 | |
| 70 | 2.68 | 0.67 | 0.5 | 1.33 | 163931 | 163931 Info | Incyte PD:1583076 | M12272 | ADH1C | 126 | 1- and 2-Methylnaphthalene degradation, Bile acid biosynthesis, Fatty acid metabolism, Glycerolipid metabolism, Glycolysis / Gluconeogenesis, Metabolism of xenobiotics by cytochrome P450, Tyrosine metabolism |
| 71 | 2.679 | 0.76 | 0.54 | 1.42 | 162099 | 162099 Info | Incyte PD:763477 | U50929 | BHMT | 635 | Glycine, serine and threonine metabolism, Methionine metabolism, misc |
| 72 | 2.669 | 0.78 | 0.63 | 1.25 | 160366 | 160366 Info | Incyte PD:260090 | AL162049 | USP10 | 9100 | |
| 73 | 2.659 | 0.99 | 0.85 | 1.17 | 169315 | 169315 Info | Incyte PD:2591494 | NM_003944 | SELENBP1 | 8991 | |
| 74 | 2.628 | 1.2 | 1.07 | 1.12 | 169441 | 169441 Info | Incyte PD:2058949 | W24219 | NDUFA1 | 4694 | Electron Transport Reaction in Mitochondria, Oxidative phosphorylation |
| 75 | 2.61 | 1.15 | 0.99 | 1.16 | 169675 | 169675 Info | Incyte PD:1215248 | NM_000154 | GALK1 | 2584 | Leloir pathway of galactose metabolism, Galactose metabolism, immunology |
| 76 | 2.576 | 0.9 | 0.77 | 1.17 | 165440 | 165440 Info | Incyte PD:1812030 | AI866254 | PBLD | 64081 | |
| 77 | 2.57 | 0.74 | 0.6 | 1.24 | 162162 | 162162 Info | Incyte PD:5035985 | M16974 | C8A | 731 | Alternative Complement Pathway, Classical Complement Pathway, Complement Pathway, Lectin Induced Complement Pathway, Complement and coagulation cascades, immunology |
| 78 | 2.549 | 0.82 | 0.68 | 1.19 | 160741 | 160741 Info | Incyte PD:2043488 | AI051566 | ALDH8A1 | 64577 | |
| 79 | 2.544 | 1.09 | 0.91 | 1.2 | 163597 | 163597 Info | Incyte PD:1755193 | AK000373 | SYBU | 55638 | |
| 80 | 2.544 | 0.99 | 0.81 | 1.22 | 167287 | 167287 Info | Incyte PD:2103752 | AI064915 | CYB5A | 1528 | |
| 81 | 2.54 | 0.96 | 0.85 | 1.13 | 162192 | 162192 Info | Incyte PD:1988940 | AA505399 | ACADM | 34 | beta-Alanine metabolism, Fatty acid metabolism, PPAR signaling pathway, Propanoate metabolism, Valine, leucine and isoleucine degradation, immunology |
| 82 | 2.505 | 1 | 0.83 | 1.21 | 168791 | 168791 Info | Incyte PD:1965695 | AF104913 | EIF4G1 | 1981 | Eukaryotic protein translation, Internal Ribosome entry pathway, mTOR Signaling Pathway, Regulation of eIF4e and p70 S6 Kinase, immunology |
| 83 | 2.473 | 0.81 | 0.65 | 1.26 | 166785 | 166785 Info | Incyte PD:778212 | AW204828 | SLC38A4 | 55089 | |
| 84 | 2.469 | 0.95 | 0.83 | 1.14 | 167390 | 167390 Info | Incyte PD:1217904 | U60873 | NEAT1 | 283131 | |
| 85 | -4.418 | 0.86 | 1.58 | 0.55 | 165127 | 165127 Info | Incyte PD:29365 | AA334619 | AFP | 174 | immunology |
| 86 | -3.559 | 1.01 | 1.24 | 0.81 | 167243 | 167243 Info | Incyte PD:387863 | NM_003097 | SNRPN | 6638 | immunology |
| 87 | -3.384 | 1.48 | 2.12 | 0.7 | 162373 | 162373 Info | Incyte PD:645584 | U50410 | GPC3 | 2719 | |
| 88 | -3.353 | 1 | 1.14 | 0.88 | 161147 | 161147 Info | Incyte PD:2416415 | NM_004760 | STK17A | 9263 | apoptosis, signal_transduction |
| 89 | -3.233 | 1.08 | 1.34 | 0.8 | 163304 | 163304 Info | Incyte PD:1420380 | U76705 | IGF2BP3 | 10643 | |
| 90 | -3.212 | 0.99 | 1.12 | 0.88 | 166900 | 166900 Info | Incyte PD:429577 | AA417813 | STEAP4 | 79689 | |
| 91 | -3.147 | 1.08 | 1.52 | 0.71 | 162399 | 162399 Info | Incyte PD:2060823 | AI151190 | S100P | 6286 | |
| 92 | -3.093 | 0.84 | 1.01 | 0.83 | 160418 | 160418 Info | Incyte PD:444957 | AB013382 | DUSP6 | 1848 | Regulation of MAP Kinase Pathways Through Dual Specificity Phosphatases, MAPK signaling pathway |
Filtering parameters:
R version 2.13.2 (2011-09-30)
Name of the project file: HCCProject.xls
Time of the analysis: Mon Jan 07 13:07:20 2013
BRB – Array Tools Version: 4.2.1 – Stable Release (January 2012)
Project annotated by SOURCE (source stanford.edu), searched by gene identifier: Symbol, on 4/30/2011 5:52:48 PM
- Apply Background adjustment: OFF
- Spot Filters: OFF
- Average the replicate spots within an array: OFF
- Use a common reference design: OFF
- Normalization: OFF
- Gene Filters: OFF
- Gene Subsets: OFF