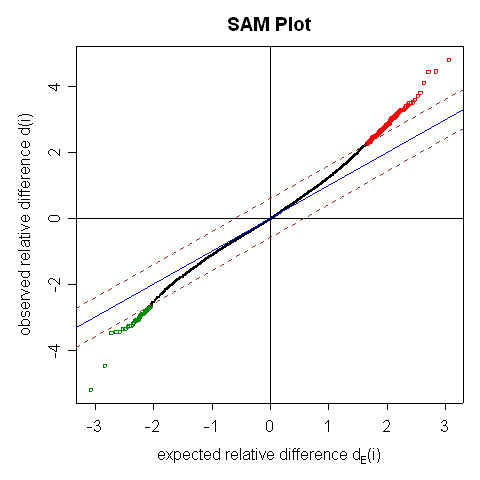
SAM (Significance Analysis of Microarray) Results
Description of the problem:
Number of classes: 2
Number of genes: 12776
Number of genes that passed filtering criteria: 8324
Type of test used: SAM – Significance analysis of Microarray
Column of the Experiment Descriptors sheet that defines class variable : GROUP
Genes which discriminate among classes:
Table 1
There are 166 genes significant by SAM
The median false discovery rate among the 166 significant genes is 0.08539
The delta value used to identify the significant genes is 0.59305
The fudge factor for standard deviation is computed as 0.03459
| Geometric mean of ratios ( Disease / Normal ) |
Qiagen oligo ID | Description | GB acc | UG cluster | Gene symbol | |
|---|---|---|---|---|---|---|
| 1 | 1.553 | H002655_01 | Collagen, type VII, alpha 1 (epidermolysis bullosa, dystrophic, dominant and recessive) | L02870 | 1640 | COL7A1 |
| 2 | 1.453 | H007688_01 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | AF109735 | 195471 | PFKFB3 |
| 3 | 1.757 | H002994_01 | Collagen, type XVIII, alpha 1 | AF018081 | 78409 | COL18A1 |
| 4 | 1.689 | H016341_01 | Platelet derived growth factor C | NM_016205 | 43080 | PDGFC |
| 5 | 1.422 | H006183_01 | Metallothionein 1X | X65607 | 278462 | MT1X |
| 6 | 1.251 | H011854_01 | Heterogeneous nuclear ribonucleoprotein C (C1/C2) |
M16342 | 182447 | HNRPC |
| 7 | 1.352 | H000498_01 | Desmoplakin (DPI, DPII) | AL031058 | 74316 | DSP |
| 8 | 1.249 | H003776_01 | Aldehyde dehydrogenase 2 family (mitochondrial) |
X05409 | 195432 | ALDH2 |
| 9 | 1.436 | H012791_01 | Metallothionein 1A (functional) | K01383 | 203967 | MT1A |
| 10 | 1.314 | H003383_01 | Ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
Z82188 | 173466 | RAC2 |
| 11 | 1.66 | H015187_01 | Homo sapiens clone N97 immunoglobulin heavy chain variable region mRNA, partial CDS |
AF103295 | 283882 | |
| 12 | 1.483 | H008423_01 | EH-domain containing 3 | NM_014600 | 87125 | EHD3 |
| 13 | 1.333 | H007009_01 | Fibroblast growth factor receptor-like 1 |
AK000530 | 193326 | FGFRL1 |
| 14 | 1.23 | H000238_01 | KRAB associated protein 1 | X97548 | 228059 | TIF1B |
| 15 | 1.386 | H011703_01 | Hypothetical protein FLJ20364 | AK000371 | 32471 | FLJ20364 |
| 16 | 1.292 | H014948_01 | Phosphatidylglycerophosphate Synthase |
NM_024419 | 278682 | PGS1 |
| 17 | 1.375 | H006492_01 | Homo sapiens CD44 isoform RC (CD44) mRNA, complete CDS |
AF098641 | 306278 | |
| 18 | 1.374 | H012664_01 | metallothionein 1B (functional) | M13485 | 211924 | MT1B |
| 19 | 1.292 | H006579_01 | Hypothetical protein FLJ10292 | AK001154 | 104650 | FLJ10292 |
| 20 | 1.427 | H008064_01 | KIAA0008 gene product | D13633 | 77695 | KIAA0008 |
| 21 | 1.255 | H012398_01 | KIAA1118 protein | AB029041 | 209646 | KIAA1118 |
| 22 | 1.451 | H003725_01 | Cartilage oligomeric matrix protein (pseudoachondroplasia, epiphyseal dysplasia 1, multiple) |
AC003107 | 1584 | COMP |
| 23 | 1.211 | H011172_01 | Heat shock protein 75 | NM_016292 | 182366 | TRAP1 |
| 24 | 1.243 | H009594_01 | Neuropathy target esterase | AJ004832 | 5038 | NTE |
| 25 | 1.445 | H008685_01 | Metallothionein 2A | V00594 | 118786 | MT2A |
| 26 | 1.279 | H007583_01 | Kinesin family member 4A | AF071592 | 279766 | KIF4A |
| 27 | 1.367 | H013251_01 | KIAA1329 protein | AB037750 | 21061 | KIAA1329 |
| 28 | 1.189 | H015295_01 | Homo sapiens HSPC337 mRNA, partial CDS |
AF161451 | 284295 | |
| 29 | 1.5 | H011653_01 | Vanilloid receptor-like protein | NM_016113 | 300686 | VRL |
| 30 | 1.405 | H012717_01 | Metallothionein 1F (functional) | M10943 | 203936 | MT1F |
| 31 | 1.193 | H003987_01 | PRO0659 protein | NM_014138 | 6451 | PRO0659 |
| 32 | 1.415 | H009923_01 | Vanilloid receptor-like protein 1 | NM_015930 | 279746 | VRL-1 |
| 33 | 1.313 | H016122_01 | TMTSP for transmembrane molecule with thrombospondin module |
NM_018676 | 325667 | LOC55901 |
| 34 | 1.244 | H012710_01 | Hepatitis delta antigen-interacting protein A |
U63825 | 66713 | DIPA |
| 35 | 0.621 | H003528_01 | Decay accelerating factor for complement (CD55, Cromer blood group system) |
M30142 | 1369 | DAF |
| 36 | 0.548 | H003827_01 | Serum/glucocorticoid regulated kinase |
AJ000512 | 296323 | SGK |
| 37 | 0.548 | H016371_01 | Hypothetical protein FLJ21212 | NM_024642 | 47099 | FLJ21212 |
| 38 | 0.584 | H009347_01 | Neuronal cell adhesion molecule | AB002341 | 7912 | NRCAM |
| 39 | 0.465 | H001509_01 | Aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) |
D17793 | 78183 | AKR1C3 |
| 40 | 0.517 | H003858_01 | Aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-a |
U05598 | 201967 | AKR1C2 |
| 41 | 0.62 | H010655_01 | Hypothetical protein FLJ20546 | AK000953 | 279896 | FLJ20546 |
| 42 | 0.811 | H005438_01 | Homo sapiens mRNA; cDNA DKFZp434B1620 (from clone DKFZp434B1620) | AL137548 | 43112 | |
| 43 | 0.728 | H004078_01 | Heme-binding protein | NM_015987 | 108675 | HEBP |
| 44 | 0.708 | H002860_01 | Inositol polyphosphate-1-phosphatase |
L08488 | 32309 | INPP1 |
| 45 | 0.768 | H002574_01 | Alcohol dehydrogenase 5 (class III), chi polypeptide |
M81118 | 78989 | ADH5 |
| 46 | 0.751 | H004767_01 | Tetraspan 3 | AK001326 | 100090 | TSPAN-3 |
| 47 | 0.629 | H010640_01 | Homo sapiens mRNA; cDNA DKFZp434B1272 (from clone DKFZp434B1272); partial CDS |
AL162032 | 158258 | |
| 48 | 0.689 | H000959_01 | Glycophorin C (Gerbich blood group) |
NM_002101 | 81994 | GYPC |
| 49 | 0.721 | H003965_01 | Cellular repressor of E1A-stimulated genes |
AF084523 | 5710 | CREG |
| 50 | 0.724 | H002462_01 | Receptor tyrosine kinase-like orphan receptor 1 |
M97675 | 274243 | ROR1 |
| 51 | 0.664 | H007733_01 | Retinal short-chain dehydrogenase/reductase retSDR3 |
NM_016246 | 18788 | LOC51171 |
| 52 | 0.771 | H016494_01 | Hypothetical protein FLJ12436 | NM_024661 | 69485 | FLJ12436 |
| 53 | 0.742 | H008089_01 | KIAA0469 gene product | AB007938 | 7764 | KIAA0469 |
| 54 | 0.631 | H007034_01 | Histamine N-methyltransferase | U44111 | 81182 | HNMT |
| 55 | 0.706 | H000410_01 | A kinase (PRKA) anchor protein (gravin) 12 |
U81607 | 788 | AKAP12 |
| 56 | 0.702 | H001356_01 | KIAA0537 gene product | AB011109 | 200598 | KIAA0537 |
| 57 | 0.538 | H001341_01 | Platelet-derived growth factor receptor-like |
D37965 | 170040 | PDGFRL |
| 58 | 0.815 | H002021_01 | V-maf musculoaponeurotic fibrosarcoma (avian) oncogene family, protein G |
AF059195 | 252229 | MAFG |
| 59 | 0.592 | H010453_01 | 3-alpha hydroxysteroid dehydrogenase type IIb |
NM_016253 | 279801 | LOC51708 |
| 60 | 0.685 | H005634_01 | Solute carrier family 11 (proton-coupled divalent metal ion transporters), member 3 |
NM_014585 | 5944 | SLC11A3 |
| 61 | 0.68 | H008745_01 | Acid sphingomyelinase-like phosphodiesterase |
AK000184 | 42945 | ASM3A |
| 62 | 0.704 | H003219_01 | Glutathione S-transferase M3 (brain) |
AF043105 | 2006 | GSTM3 |
| 63 | 0.773 | H003951_01 | Predicted osteoblast protein | D87120 | 29882 | GS3786 |
| 64 | 0.735 | H001047_01 | Nidogen 2 | AB009799 | 82733 | NID2 |
| 65 | 0.792 | H002382_01 | Limbic system-associated membrane protein |
U41901 | 26479 | LSAMP |
| 66 | 0.623 | H016583_01 | 1,2-alpha-mannosidase IC | NM_020379 | 8910 | HMIC |
| 67 | 0.742 | H011191_01 | Lipin 2 | D87436 | 166318 | LPIN2 |
| 68 | 0.803 | H002185_01 | Niemann-Pick disease, type C1 | AF002020 | 76918 | NPC1 |
| 69 | 0.775 | H000509_01 | Neural precursor cell expressed, developmentally down-regulated 4 |
D42055 | 1565 | NEDD4 |
| 70 | 0.786 | H007419_01 | Progesterone membrane binding protein |
AJ002030 | 9071 | PMBP |
| 71 | 0.774 | H002121_01 | Paraoxonase 2 | AF001601 | 169857 | PON2 |
| 72 | 0.805 | H008087_01 | HSPC019 protein | NM_014028 | 163724 | HSPC019 |
| 73 | 0.696 | H002393_01 | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
AB000220 | 171921 | SEMA3C |
| 74 | 0.785 | H014975_01 | Homo sapiens mRNA; cDNA DKFZp564N1272 (from clone DKFZp564N1272); complete CDS |
AF087847 | 282654 | |
| 75 | 0.614 | H001215_01 | Pentaxin-related gene, rapidly induced by IL-1 beta |
M31166 | 2050 | PTX3 |
| 76 | 0.756 | H002423_01 | Vascular endothelial growth factor B |
U43368 | 78781 | VEGFB |
| 77 | 0.768 | H010087_01 | Destrin (actin depolymerizing factor) |
S65738 | 82306 | ADF |
| 78 | 0.742 | H000025_01 | Neutral sphingomyelinase (N-SMase) activation associated factor |
X96586 | 78687 | NSMAF |
| 79 | 0.744 | H005560_01 | WAS protein family, member 1 | D87459 | 75850 | WASF1 |
| 80 | 0.786 | H014051_01 | Very long-chain acyl-CoA synthetase homolog 2 |
NM_012254 | 111401 | VLCS-H2 |
| 81 | 0.644 | H004741_01 | Fibroblast growth factor 13 | AF100143 | 6540 | FGF13 |
| 82 | 0.764 | H001605_01 | Annexin A4 | NM_001153 | 77840 | ANXA4 |
| 83 | 0.71 | H016019_01 | Duodenal cytochrome b | AL136693 | 31297 | FLJ23462 |
| 84 | 0.804 | H001346_01 | Inositol(myo)-1(or 4)-monophosphatase 2 |
NM_014214 | 5753 | IMPA2 |
| 85 | 0.635 | H010918_01 | WNT1 inducible signaling pathway protein 2 |
AF100780 | 194679 | WISP2 |
| 86 | 0.791 | H001101_01 | CD164 antigen, sialomucin | D14043 | 43910 | CD164 |
| 87 | 0.787 | H004431_01 | Rag C protein | AF131773 | 110950 | GTR2 |
| 88 | 0.792 | H012626_01 | Beta-2-microglobulin | AB021288 | 75415 | B2M |
| 89 | 0.805 | H002819_01 | Protective protein for beta-galactosidase (galactosialidosis) |
AL008726 | 118126 | PPGB |
| 90 | 0.684 | H002497_01 | Complement component 1, r subcomponent |
M14058 | 1279 | C1R |
| 91 | 0.691 | H006676_01 | Lung type-I cell membrane-associated glycoprotein |
AF030428 | 135150 | T1A-2 |
| 92 | 0.653 | H012777_01 | KIAA1199 protein | AB033025 | 50081 | KIAA1199 |
| 93 | 0.721 | H003789_01 | Colony stimulating factor 1 (macrophage) |
M37435 | 173894 | CSF1 |
| 94 | 0.849 | H008439_01 | SCO (cytochrome oxidase deficient, yeast) homolog 1 |
AF026852 | 14511 | SCO1 |
| 95 | 0.774 | H008994_01 | Solute carrier family 31 (copper transporters), member 2 |
U83461 | 24030 | SLC31A2 |
| 96 | 0.774 | H010497_01 | Interferon induced transmembrane protein 3 (1-8U) |
X57352 | 182241 | IFITM3 |
| 97 | 0.762 | H011175_01 | Opsin 3 (encephalopsin) | NM_014322 | 279926 | OPN3 |
| 98 | 0.701 | H011671_01 | Short-chain dehydrogenase/reductase 1 |
AF061741 | 17144 | SDR1 |
| 99 | 0.767 | H002804_01 | Prion protein (p27-30) (Creutzfeldt-Jakob disease, Gerstmann-Strausler-Scheinker syndrome, fatal fami |
U29185 | 74621 | PRNP |
| 100 | 0.792 | H002258_01 | UDP-glucose pyrophosphorylase 2 | U27460 | 77837 | UGP2 |
| 101 | 0.659 | H001491_01 | Cyclin T2 | AF048731 | 155478 | CCNT2 |
| 102 | 0.762 | H002746_01 | TNF receptor-associated factor 5 | AB000509 | 29736 | TRAF5 |
| 103 | 0.766 | H013070_01 | Sarcoglycan, epsilon | AJ000534 | 110708 | SGCE |
| 104 | 0.761 | H001062_01 | N-acylsphingosine amidohydrolase (acid ceramidase) |
U70063 | 75811 | ASAH |
| 105 | 0.814 | H002694_01 | Glutathione peroxidase 4 (phospholipid hydroperoxidase) |
X71973 | 2706 | GPX4 |
| 106 | 0.774 | H011917_01 | Sorting nexin 3 | AF034546 | 12102 | SNX3 |
| 107 | 0.754 | H006813_01 | Homo sapiens cDNA FLJ11309 fis, clone PLACE1010076 |
AK002171 | 28005 | |
| 108 | 0.821 | H002793_01 | Antigen identified by monoclonal antibodies 12E7, F21 and O13 |
M16279 | 177543 | MIC2 |
| 109 | 0.844 | H000474_01 | Testis-specific kinase 1 | D50863 | 79358 | TESK1 |
| 110 | 0.731 | H000930_01 | Low density lipoprotein receptor (familial hypercholesterolemia) |
L00352 | 213289 | LDLR |
| 111 | 0.802 | H009135_01 | Electron-transferring-flavoprotein dehydrogenase |
S69232 | 323468 | ETFDH |
| 112 | 0.767 | H008544_01 | Atrophin-1 interacting protein 1; activin receptor interacting protein 1 |
AB014605 | 22599 | KIAA0705 |
| 113 | 0.789 | H000400_01 | Growth arrest-specific 6 | L13720 | 78501 | GAS6 |
| 114 | 0.762 | H016469_01 | MEK partner 1 | NM_021970 | 6361 | MP1 |
| 115 | 0.731 | H002820_01 | Ectodermal-neural cortex (with BTB-like domain) |
AF059611 | 104925 | ENC1 |
| 116 | 0.75 | H016592_01 | Hypothetical protein FLJ12816 | NM_022060 | 9175 | FLJ12816 |
| 117 | 0.792 | H006873_01 | CGI-149 protein | NM_016079 | 189658 | LOC51652 |
| 118 | 0.592 | H002066_01 | Aldehyde dehydrogenase 1 family, member A1 |
M31994 | 76392 | ALDH1A1 |
| 119 | 0.689 | H013490_01 | H.sapiens polyA site DNA | Z24724 | 4934 | |
| 120 | 0.76 | H015305_01 | Hypothetical protein FLJ22029 | NM_024949 | 285243 | FLJ22029 |
| 121 | 0.704 | H002566_01 | Membrane metallo-endopeptidase (neutral endopeptidase, enkephalinase, CALLA, CD10) |
NM_007289 | 1298 | MME |
| 122 | 0.81 | H002169_01 | Sortilin-related receptor, L(DLR class) A repeats-containing |
NM_003105 | 278571 | SORL1 |
| 123 | 0.88 | H003649_01 | Protein tyrosine phosphatase, non-receptor type 6 |
X62055 | 63489 | PTPN6 |
| 124 | 0.744 | H011942_01 | Activating transcription factor 5 | AB021663 | 9754 | ATF5 |
| 125 | 0.806 | H007715_01 | KIAA1091 protein | AL117448 | 26797 | KIAA1091 |
| 126 | 0.823 | H012019_01 | Epithelial protein lost in neoplasm beta |
NM_016357 | 10706 | EPLIN |
| 127 | 0.838 | H010079_01 | Hypothetical protein FLJ10337 | AB037858 | 173484 | KIAA1437 |
| 128 | 0.772 | H003031_01 | Glycoprotein A repetitions predominant |
Z24680 | 151641 | GARP |
| 129 | 0.762 | H002785_01 | Serine (or cysteine) proteinase inhibitor, clade G (C1 inhibitor), member 1 |
X54486 | 151242 | SERPING1 |
| 130 | 0.631 | H000082_01 | Deiodinase, iodothyronine, type II | NM_013989 | 154424 | DIO2 |
| 131 | 0.755 | H007381_01 | Hypothetical protein FLJ10044 | AK000906 | 127273 | FLJ10044 |
| 132 | 0.699 | H014982_01 | Tuftelin 1 | NM_020127 | 283009 | TUFT1 |
| 133 | 0.813 | H003637_01 | Protein tyrosine phosphatase, non-receptor type 12 |
M93425 | 62 | PTPN12 |
| 134 | 0.821 | H002684_01 | Bone morphogenetic protein 6 | NM_001718 | 285671 | BMP6 |
| 135 | 0.847 | H001078_01 | ATPase, Na+/K+ transporting, beta 3 polypeptide |
NM_001679 | 76941 | ATP1B3 |
| 136 | 0.785 | H009413_01 | KIAA0321 protein | AB002319 | 8663 | KIAA0321 |
| 137 | 0.806 | H003439_01 | Protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform |
M60484 | 80350 | PPP2CB |
| 138 | 0.811 | H011488_01 | UDP-Gal:beta GlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
AL035683 | 107526 | B4GALT5 |
| 139 | 0.631 | H002334_01 | Cytochrome P450, subfamily I (dioxin-inducible), polypeptide 1 (glaucoma 3, primary infantile) |
U03688 | 154654 | CYP1B1 |
| 140 | 0.797 | H006973_01 | Homo sapiens cDNA FLJ10598 fis, clone NT2RP2004841 |
AK001460 | 170873 | |
| 141 | 0.737 | H008497_01 | Amyloid beta (A4) precursor-like protein 2 |
NM_016160 | 279518 | APLP2 |
| 142 | 0.802 | H000517_01 | Phorbol-12-myristate-13-acetate-induced protein 1 |
D90070 | 96 | PMAIP1 |
| 143 | 0.799 | H007506_01 | Clock (mouse) homolog | AB002332 | 50722 | CLOCK |
| 144 | 0.831 | H002305_01 | Accessory proteins BAP31/BAP29 | S74039 | 291904 | DXS1357E |
| 145 | 0.813 | H002571_01 | Meiotic recombination (S. cerevisiae) 11 homolog B |
AF022778 | 202379 | MRE11B |
| 146 | 0.795 | H002782_01 | Wee1+ (S. pombe) homolog | X62048 | 75188 | WEE1 |
| 147 | 0.814 | H002076_01 | Interleukin-1 receptor-associated kinase 1 |
L76191 | 182018 | IRAK1 |
| 148 | 0.82 | H000048_01 | PRKC, apoptosis, WT1, regulator | U63809 | 176090 | PAWR |
| 149 | 0.578 | H005801_01 | Purkinje cell protein 4 | U52969 | 80296 | PCP4 |
| 150 | 0.834 | H002311_01 | Transcription factor 7-like 2 (T-cell specific, HMG-box) |
Y11306 | 173638 | TCF7L2 |
| 151 | 0.774 | H008367_01 | KIAA0623 gene product | AB014523 | 151406 | KIAA0623 |
| 152 | 0.809 | H005443_01 | Interferon-related developmental regulator 1 |
AC005192 | 7879 | IFRD1 |
| 153 | 0.796 | H005941_01 | Hypothetical protein FLJ20043 | AK000050 | 103853 | FLJ20043 |
| 154 | 0.731 | H009185_01 | KIAA1144 protein | AB032970 | 22675 | KIAA1144 |
| 155 | 0.737 | H011708_01 | Homo sapiens clone 24405 mRNA sequence |
AF070632 | 23729 | |
| 156 | 0.848 | H016365_01 | Core1 UDP-galactose:N-acetylgalactosamine-alpha-R beta 1,3-galactosyltransferase |
NM_020156 | 46744 | C1GALT1 |
| 157 | 0.763 | H010122_01 | Homo sapiens mRNA; cDNA DKFZp586F1223 (from clone DKFZp586F1223) |
AL050204 | 28540 | |
| 158 | 0.71 | H002475_01 | Pim-1 oncogene | M54915 | 81170 | PIM1 |
| 159 | 0.794 | H007334_01 | SH3 domain binding glutamic acid-rich protein like |
AF042081 | 14368 | SH3BGRL |
| 160 | 0.817 | H000893_01 | Major histocompatibility complex, class I, E |
X56841 | 181392 | HLA-E |
| 161 | 0.737 | H010951_01 | LIM domain only 4 | NM_006769 | 3844 | LMO4 |
| 162 | 0.846 | H001759_01 | Chloride channel 7 | AL031600 | 80768 | CLCN7 |
| 163 | 0.845 | H001612_01 | RAP1, GTP-GDP dissociation stimulator 1 |
AF237413 | 7940 | RAP1GDS1 |
| 164 | 0.753 | H007858_01 | Glutaminyl-peptide cyclotransferase (glutaminyl cyclase) |
X71125 | 79033 | QPCT |
| 165 | 0.808 | H002897_01 | Cathepsin L | X12451 | 78056 | CTSL |
| 166 | 0.84 | H004099_01 | Phosphatidic acid phosphatase type 2A |
AF014403 | 41569 | PPAP2A |
‘Observed v. Expected’ table of GO classes and parent classes, in list of 166 genes shown above:
Only GO classes and parent classes with at least 5 observations in the selected subset and with an ‘Observed vs. Expected’ ratio of at least 2 are shown.
Cellular Component
| GO id | GO classification | Observed in selected subset |
Expected in selected subset |
Observed/ Expected |
|---|---|---|---|---|
| 0005887 | integral to plasma membrane | 19 | 9.37 | 2.03 |
Molecular Function
| GO id | GO classification | Observed in selected subset |
Expected in selected subset |
Observed/ Expected |
|---|---|---|---|---|
| 0016791 | phosphoric monoester hydrolase activity | 6 | 2.04 | 2.94 |
| 0005489 | electron transporter activity | 7 | 2.54 | 2.76 |
| 0042578 | phosphoric ester hydrolase activity | 6 | 2.4 | 2.5 |
| 0004713 | protein-tyrosine kinase activity | 5 | 2.16 | 2.31 |
Biological Process
| GO id | GO classification | Observed in selected subset |
Expected in selected subset |
Observed/ Expected |
|---|---|---|---|---|
| 0007600 | sensory perception | 6 | 1.92 | 3.13 |
| 0009581 | detection of external stimulus | 6 | 2.13 | 2.82 |
| 0006812 | cation transport | 6 | 2.93 | 2.05 |