Supplement 2: Hierarchical Clustering of the transcripts differentially expressed in SLE and SSc in comparison to controls
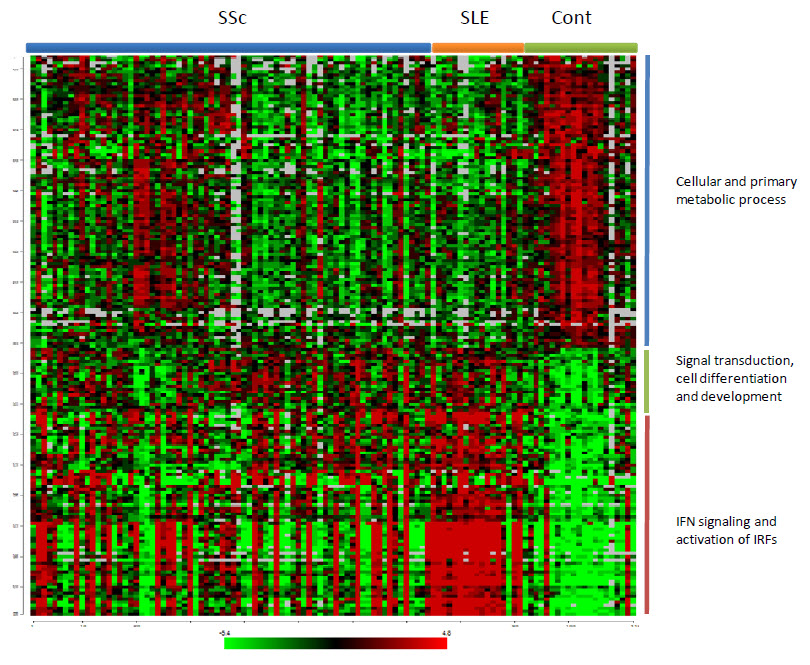
These are the major categories of the 185 differentially expressed genes that were in common between SSc and SLE (Figure 1). The upper two sets of transcripts were broadly grouped according to their gene ontology biologic process using DAVID or Database for Annotation Visualization and Integrated Discovery.
The genes belonging to the IFN signaling and activation of IRFs pathways were overexpressed in the majority of SLE patients and in a group of patients with SSc. We observed a similar picture among the genes belonging to the signal transduction, cell differentiation and development. The genes belonging to the cellular and primary metabolic process were underexpressed in the majority of SLE patients and in a group of patients with SSc.