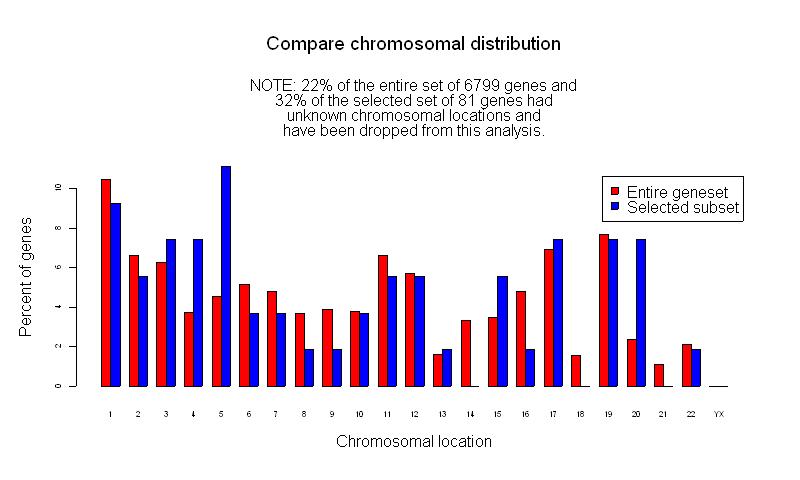
SAM (Significance Analysis of Microarray) Results
Description of the problem:
Number of classes: 2
Number of genes: 16657
Number of genes that passed filtering criteria: 6799
Type of test used: SAM – Significance analysis of Microarray
Column of the Experiment Descriptors sheet that defines class variable : All SSc affected twins vs population controls and healthy DZ twin
Genes which discriminate among classes:
Table 1
There are 81 genes significant by SAM
The median false discovery rate among the 81 significant genes is 0.09328.
The delta value used to identify the significant genes is 0.57426.
The fudge factor for standard deviation is computed as 0.08062.
Note: For the latest annotations please query the Qiagen website using the Oligo ID number.
| Geom mean of ratios Normal |
Geom mean of ratios in SSc |
Fold difference of geom means |
Qiagen Oligo ID | Description | GB acc | UG cluster | Gene symbol | |
| 15 | 2.554 | 14.186 | 5.55 | H001924_01 | Thy-1 cell surface antigen |
AL161958 | 125359 | THY1 |
| 20 | 1.849 | 10.168 | 5.50 | H003853_01 | Activin A receptor, type IB |
Z22536 | 99954 | ACVR1B |
| 21 | 3.098 | 14.225 | 4.59 | H002800_01 | Connective tissue growth factor |
X78947 | 75511 | CTGF |
| 45 | 2.813 | 12.316 | 4.38 | H000899_01 | UDP glycosyltransferase 2 family, polypeptide B7 |
J05428 | 10319 | UGT2B7 |
| 50 | 3.473 | 15.156 | 4.36 | H005487_01 | RHCE | S70456 | 251513 | |
| 36 | 1.753 | 7.48 | 4.27 | H007489_01 | KIAA0943 protein |
AB023160 | 272586 | KIAA0943 |
| 27 | 2.355 | 9.975 | 4.24 | H013988_01 | Hypothetical protein FLJ21347 |
NM_022827 | 103147 | FLJ21347 |
| 58 | 3.691 | 14.522 | 3.93 | H000555_01 | MAD2 (mitotic arrest deficient, yeast, homolog)-like 1 |
U65410 | 79078 | MAD2L1 |
| 38 | 0.321 | 1.23 | 3.83 | H008524_01 | Angiotensin receptor-like 2 |
NM_005162 | 279535 | AGTRL2 |
| 52 | 1.736 | 6.615 | 3.81 | H009493_01 | Rhesus blood group, CcEe antigens |
NM_016191 | 278994 | RHCE |
| 61 | 2.633 | 9.908 | 3.76 | H004056_01 | Murine retrovirus integration site 1 homolog |
NM_006069 | 251385 | MRVI1 |
| 23 | 3.259 | 12.005 | 3.68 | H003218_01 | Calcium channel, voltage-dependent, L type, alpha 1D subunit |
M76558 | 23838 | CACNA1D |
| 39 | 3.646 | 13.226 | 3.63 | H006619_01 | Hypothetical protein FLJ20393 |
AK000400 | 326545 | FLJ20393 |
| 43 | 3.38 | 12.07 | 3.57 | H005913_01 | Apical protein, Xenopus laevis like |
X83543 | 2391 | APXL |
| 48 | 2.295 | 8.18 | 3.56 | H012790_01 | Phosphatidylinositol 4-kinase type II |
AF070611 | 25300 | PI4KII |
| 56 | 3.178 | 11.31 | 3.56 | H012306_01 | Carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
AL022165 | 138155 | CHST7 |
| 22 | 1.259 | 4.463 | 3.54 | H010437_01 | NY-REN-24 antigen |
AF052087 | 128425 | LOC58509 |
| 80 | 1.863 | 6.474 | 3.48 | H010823_01 | Homo sapiens cDNA FLJ11154 fis, clone PLACE1006932 |
AK002016 | 114727 | |
| 35 | 2.917 | 9.772 | 3.35 | H003072_01 | Collagen, type I, alpha 2 |
J03464 | 179573 | COL1A2 |
| 33 | 3.568 | 11.929 | 3.34 | H004727_01 | Putative transmembrane protein; homolog of yeast Golgi membrane protein Yif1p (Yip1p-interacting fac |
AF004876 | 5809 | 54TM |
| 18 | 2.45 | 8.168 | 3.33 | H003695_01 | Secreted protein, acidic, cysteine-rich (osteonectin) |
J03040 | 111779 | SPARC |
| 42 | 1.585 | 5.146 | 3.25 | H000408_01 | Branched chain aminotransferase 2, mitochondrial |
U68418 | 101408 | BCAT2 |
| 59 | 1.005 | 3.139 | 3.12 | H005176_01 | H3 histone family, member I |
Z80786 | 247814 | H3FI |
| 67 | 0.988 | 2.893 | 2.93 | H003699_01 | Chondroitin sulfate proteoglycan 2 (versican) |
U16306 | 81800 | CSPG2 |
| 46 | 0.637 | 1.85 | 2.90 | H002468_01 | Homo sapiens inducible nitric oxide synthase (iNOS) mRNA, complete cds |
AF049656 | 248084 | |
| 40 | 1.45 | 4.056 | 2.80 | H009480_01 | Tropomyosin 1 (alpha) |
AL050179 | 77899 | TPM1 |
| 76 | 5.997 | 16.427 | 2.74 | H010386_01 | Human DNA sequence from clone RP5-1103G7 on chromosome 20p12.2-13. Contains up to three novel genes, |
AL034548 | 96151 | |
| 71 | 1.699 | 4.652 | 2.74 | H002915_01 | Collagen, type VIII, alpha 1 |
X57527 | 114599 | COL8A1 |
| 28 | 1.714 | 4.608 | 2.69 | H002144_01 | Serine (or cysteine) proteinase inhibitor, clade H (heat shock protein 47), member 1 |
X61598 | 241579 | SERPINH1 |
| 41 | 0.828 | 2.145 | 2.59 | H008112_01 | TATA box binding protein (TBP)-associated factor, RNA polymerase II, J, 20kD |
U57693 | 82037 | TAF2J |
| 17 | 0.658 | 1.685 | 2.56 | H015129_01 | Gap junction protein, alpha 3, 46kD (connexin 46) |
NM_021954 | 283746 | GJA3 |
| 30 | 0.77 | 1.942 | 2.52 | H015287_01 | Homo sapiens HSPC080 mRNA, partial cds |
AF161343 | 284278 | |
| 29 | 0.785 | 1.947 | 2.48 | H004244_01 | 75 kda infertility-related sperm protein [human, testis, mRNA Partial, 2427 nt] |
S58544 | 62608 | |
| 77 | 2.326 | 5.749 | 2.47 | H010247_01 | ADP-ribosylation factor related protein 1 |
X91504 | 64904 | ARFRP1 |
| 31 | 1.248 | 3.002 | 2.41 | H008600_01 | Paralemmin | Y16270 | 78482 | PALM |
| 44 | 0.58 | 1.377 | 2.37 | H014899_01 | Hypothetical protein FLJ13346 |
NM_024918 | 266273 | FLJ13346 |
| 70 | 0.97 | 2.281 | 2.35 | H005180_01 | Tubby like protein 3 |
AF045583 | 169084 | TULP3 |
| 25 | 0.637 | 1.488 | 2.34 | H002448_01 | Fibroblast growth factor receptor 3 (achondroplasia, thanatophoric dwarfism) |
M64347 | 1420 | FGFR3 |
| 81 | 1.321 | 3.084 | 2.33 | H001268_01 | ELL-RELATED RNA POLYMERASE II, ELONGATION FACTOR |
U88629 | 173334 | ELL2 |
| 16 | 1.042 | 2.336 | 2.24 | H014431_01 | Hypothetical protein FLJ12876 |
NM_022754 | 16131 | FLJ12876 |
| 53 | 1.529 | 3.303 | 2.16 | H008340_01 | Hypothetical protein 24636 |
AF070643 | 4187 | LOC55977 |
| 47 | 0.746 | 1.608 | 2.16 | H006032_01 | Homo sapiens clone 24418 mRNA sequence |
AF052185 | 13434 | |
| 62 | 0.626 | 1.338 | 2.14 | H001113_01 | Calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
M76559 | 1295 | CACNA2D1 |
| 64 | 0.827 | 1.746 | 2.11 | H000456_01 | Adenylate kinase 3 |
X60673 | 274691 | AK3 |
| 73 | 1.229 | 2.577 | 2.10 | H000277_01 | Four and a half LIM domains 2 |
L42176 | 8302 | FHL2 |
| 26 | 0.18 | 0.375 | 2.08 | H007495_01 | H1 histone family, member 2 |
X57129 | 7644 | H1F2 |
| 60 | 0.949 | 1.912 | 2.01 | H000168_01 | NGFI-A binding protein 1 (ERG1 binding protein 1) |
AF045451 | 107474 | NAB1 |
| 19 | 0.609 | 1.222 | 2.01 | H013132_01 | Human DNA sequence from clone RP3-329A5 on chromosome 6p21.1-21.33 Contains a pseudogene similar to |
Z97832 | 272289 | |
| 65 | 0.911 | 1.784 | 1.96 | H004028_01 | clg01 protein | AL021578 | 251374 | CLG01 |
| 51 | 1.98 | 3.86 | 1.95 | H009379_01 | Annexin A2 pseudogene 2 |
M62895 | 166072 | ANXA2P2 |
| 32 | 0.89 | 1.725 | 1.94 | H014653_01 | Ras homolog gene family, member B |
NM_004040 | 204354 | ARHB |
| 63 | 0.606 | 1.174 | 1.94 | H001752_01 | Ran GTPase activating protein 1 |
X82260 | 183800 | RANGAP1 |
| 54 | 0.521 | 1.003 | 1.93 | H002024_01 | Cadherin 3, type 1, P-cadherin (placental) |
X63629 | 2877 | CDH3 |
| 37 | 0.694 | 1.31 | 1.89 | H001445_01 | A disintegrin and metalloproteinase domain 19 (meltrin beta) |
AL049415 | 278679 | ADAM19 |
| 66 | 0.687 | 1.295 | 1.89 | H010888_01 | High-mobility group 20B |
AF072836 | 32317 | HMG20B |
| 24 | 0.504 | 0.937 | 1.86 | H000365_01 | Zinc finger protein 43 (HTF6) |
X59244 | 74107 | ZNF43 |
| 49 | 1.382 | 2.496 | 1.81 | H016173_01 | Homo sapiens HSPC134 protein (HSPC134), mRNA |
XM_007306 | 326630 | |
| 34 | 0.706 | 1.257 | 1.78 | H008649_01 | Parathymosin | M24398 | 171814 | PTMS |
| 57 | 1.101 | 1.942 | 1.76 | H011917_01 | Sorting nexin 3 | AF034546 | 12102 | SNX3 |
| 69 | 0.864 | 1.509 | 1.75 | H014475_01 | Mitochondrial uncoupling protein 1 |
NM_021734 | 16786 | MUP1 |
| 72 | 0.994 | 1.735 | 1.75 | H003496_01 | Small inducible cytokine subfamily B (Cys-X-Cys), member 10 |
X02530 | 2248 | SCYB10 |
| 55 | 3.978 | 6.902 | 1.74 | H001149_01 | Microfibrillar-associated protein 2 |
U19718 | 83551 | MFAP2 |
| 74 | 0.6 | 1.039 | 1.73 | H000192_01 | Putative mitochondrial outer membrane protein import receptor |
AF026031 | 31334 | TOM |
| 79 | 0.792 | 1.36 | 1.72 | H007580_01 | Homo sapiens mRNA; cDNA DKFZp761G151 (from clone DKFZp761G151); partial cds |
AL157475 | 35453 | |
| 78 | 0.661 | 1.133 | 1.71 | H004407_01 | SH3 protein | AF178432 | 102929 | AF3P21 |
| 75 | 0.341 | 0.537 | 1.57 | H004497_01 | KIAA0116 protein |
D29958 | 182877 | KIAA0116 |
| 68 | 0.536 | 0.779 | 1.45 | H006665_01 | KIAA1081 protein |
AB029004 | 293705 | KIAA1081 |
| 8 | 1.387 | 0.734 | 0.53 | H006223_01 | Hypothetical protein |
AJ012409 | 8170 | YR-29 |
| 5 | 2.149 | 1.093 | 0.51 | H001615_01 | Calmodulin 3 (phosphorylase kinase, delta) |
X52606 | 141011 | CALM3 |
| 9 | 0.738 | 0.374 | 0.51 | H012558_01 | H2A histone family, member G |
Z80776 | 239458 | H2AFG |
| 12 | 1.873 | 0.946 | 0.51 | H007570_01 | KIAA0171 gene product |
D79993 | 155623 | KIAA0171 |
| 10 | 1.692 | 0.843 | 0.50 | H002764_01 | Lipase, hepatic | X07228 | 9994 | LIPC |
| 11 | 1.079 | 0.526 | 0.49 | H009598_01 | Sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
L19956 | 274614 | SULT1A3 |
| 4 | 1.236 | 0.572 | 0.46 | H009251_01 | Splicing factor proline/glutamine rich (polypyrimidine tract-binding protein-associated) |
X70944 | 180610 | SFPQ |
| 13 | 0.858 | 0.391 | 0.46 | H013222_01 | DKFZP564I122 protein |
AL080062 | 13024 | DKFZP564I122 |
| 14 | 1.96 | 0.863 | 0.44 | H011937_01 | Dimethylarginine dimethylaminohydrolase 2 |
AJ012008 | 247362 | DDAH2 |
| 1 | 1.757 | 0.763 | 0.43 | H015502_01 | Hypothetical protein FLJ22242 |
NM_025070 | 288057 | FLJ22242 |
| 6 | 1.197 | 0.445 | 0.37 | H007811_01 | Serologically defined colon cancer antigen 3 |
AK001296 | 94300 | SDCCAG3 |
| 7 | 10.506 | 3.254 | 0.31 | H002995_01 | Apolipoprotein D |
J02611 | 75736 | APOD |
| 2 | 0.68 | 0.159 | 0.23 | H007802_01 | CGI-134 protein | AK002008 | 3385 | LOC51023 |
| 3 | 0.245 | 0.05 | 0.20 | H006006_01 | Human Ig rearranged lambda-chain mRNA, subgroup VL3, V-J region, partial cds |
L21961 | 247947 |
Observed v. Expected’ table of GO classes and parent classes, in list of 81 genes shown above:
Only GO classes and parent classes with at least 5 observations in the selected subset and with an ‘Observed vs. Expected’ ratio of at least 2 are shown.
Cellular Component
| GO id | GO classification | Observed in | Expected in | Observed/ | |
| selected subset | selected subset | Expected | |||
| 31012 | extracellular matrix |
6 | 1.12 | 5.38 | |
| 5578 | extracellular matrix (sensu Metazoa) |
6 | 1.12 | 5.38 | |
| 5576 | extracellular region |
8 | 3.19 | 2.5 | |
| 267 | cell fraction | 6 | 2.99 | 2 |
Molecular Function
| GO id | GO classification | Observed in selected subset | Expected in selected subset | Observed/Expected |
| 5198 | structural molecule activity | 5 | 2.35 | 2.13 |
Biological Process
| GO id | GO classification | Observed in selected subset | Expected in selected subset | Observed/Expected | |
| 6811 | ion transport | 5 | 1.81 | 2.76 | |
| 48513 | organ development | 6 | 2.78 | 2.16 | |
| 9887 | organogenesis | 6 | 2.78 | 2.16 |