Xenopus kidney research published in Kidney International
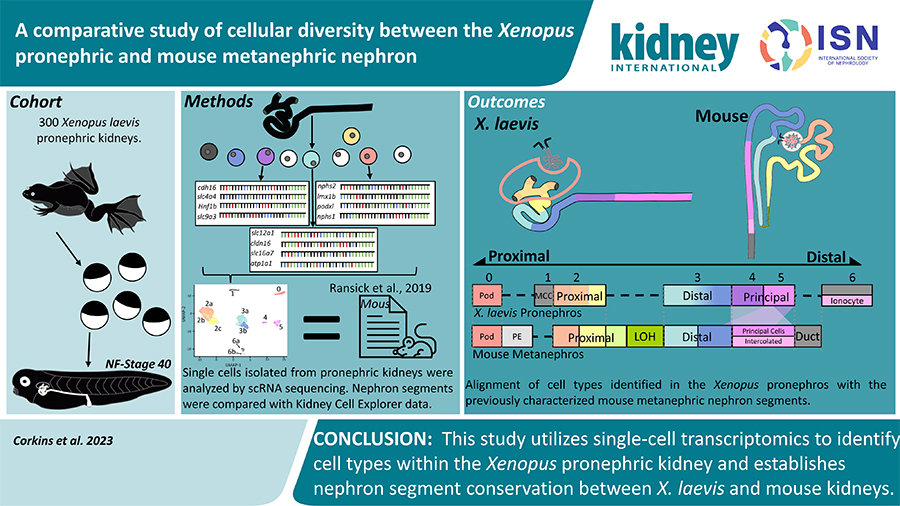

Recent research from the lab of Rachel Miller, PhD, associate professor in the Department of Pediatrics, seeking to create a better understanding of the gene expression and physiology of the Xenopus kidney, has been published in the journal Kidney International.
The Xenopus model has become a valuable tool for kidney research, offering insights into human kidney development, eliminating the need to perform experiment on humans. The Xenopus model needs just four days to form a functional kidney, and hundreds of models can be utilized in a single experiment. Despite the usefulness of the Xenopus model, a clear understanding of the similarities between mammalian and amphibian kidneys was needed.

“Xenopus has provided many novel insights into kidney development. However, looking at individual genes has led to competing models of how the Xenopus kidney compares to mammalian kidneys,” said Mark Corkins, PhD, lead author for the paper and a former postdoctoral fellow in the Miller Lab and associate scientist at Mount Sanai in New York City. “The goal of this work is to use new technologies, along with the previous efforts of other Xenopus labs to look at thousands of genes to fill in some of these gaps to get a better understanding of the underlying gene expression and physiology of the Xenopus kidney.”
Taking advantage of the ability to produce an abundance of Xenopus embryos at once, the lab was able to isolate hundreds of kidneys. Using these kidneys, the team then performed gene expression analysis on over 10,000 individual kidney cells.
After evaluating the single-cell gene expression, Corkins and the team collaborated with the lab of Dr. Nils Lindstrom, who had completed similar datasets comparing human and murine models. Using this mammalian dataset, the lab was able to identify structures that were both analogous or missing in the Xenopus kidney.
“Using this data, we were able to identify the cell types present in the Xenopus kidney to refine the previous models proposed,” Corkins said. “Given the sequencing depth and number of cells analyzed, we have high confidence that we have identified the true model of the Xenopus kidney. We are able to identify gene expression for thousands of genes and will be able to better predict which parts of the kidney human mutations are more likely to affect.”
Moving forward, the data will become a reference for future experiments in kidney research and is freely available for other researchers to use in their own projects. Specifically for the Miller Lab, there is an interest in how human diseases such as DYRK1A related intellectual disability, Down syndrome, and Li-Fraumeni syndrome lead to congenital kidney abnormalities.
Contributing authors for the story are Corkins; MaryAnne Achieng; Bridget DeLay, PhD; Vanja Krneta-Stankic, PhD; Margo Cain, PhD; Brandy Walker; Jichao Chen, PhD; Lindstrom; and Miller.