Accuracy
The Accuracy subdirectory contains simulations that were designed to test the accuracy of the SNNAP software.
The accuracy of the SNNAP software can be tested, in part, by implementing previously published models and comparing the results from SNNAP simulations to published results, which were obtained by various different methods.
This subdirectory contains two simulations. One simulation (\Epstein_Marder) is of a bursting neuron. This model was originally developed by Epstein and Marder (1990). The second simulation (\Segev_Burke) is of a branched neuron. This model was originally developed by Segev et al. 1989. In addition to these two simulations, the user can refer to the SNNAP simulations of the Hodgkin and Huxley model of the squid giant axon.
Epstein Marder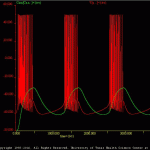
The goal of the present simulation is to implement a model published by Epstein & Marder (1990) and check the accuracy of SNNAP by comparing SNNAP – generated results with previously published results.
This subdirectory contains a model originally published by:
Epstein, I.R. and Marder, E. (1990) Multiple modes of a conditional neural oscillator. Biol. Cybern. 63: 25-34.
The present model corresponds to the “Model N” of Epstein and Marder (1990). The components of the modeled bursting neuron are illustrated in Epstein_Example.gif. The default simulation is similar to Fig. 5 of Epstein and Marder (1990).
Segev Burke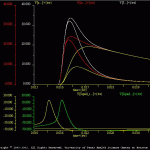
The goal of the present simulation is to implement the model of a branched neuronal structure that originally developed by Segev et al. (1989), and compare the results produced by SNNAP to the previously published results.
This example illustrates SNNAP simulations of a model that was originally developed by:
Segev, I., Fleshman, J.W, and Burke, R.E. (1989) Compartmental models of complex neurons. In: C. Koch & I. Segev (eds) Methods in Neuronal Modeling: From Synapses to Networks (pp.63-96). Cambridge: The MIT Press.
The structure of the SNNAP model is illustrated in Segev_ntw.jpg. In this model, there is a postsynaptic neuron and two presynaptic neurons. The presynaptic neurons (squid.neu and squid_i.neu) mediate excitatory (i.e., exec.cs) and inhibitory (inhib.cs) inputs to the postsynaptic neuron.
The results of the default simulation are illustrated in Segev_ous.jpg. In the default simulation, voltages in the postsynaptic neuron are monitored in compartments a, f and m. An inhibitory input to the postsynaptic neuron (i.e., spike in Squid_i.neu) immediately precedes an excitatory input (i.e., spike in Squid.neu). As a result of this inhibition, the amplitude and time course of the EPSPs is altered. To compare the EPSPs with and without inhibition, the user can use the ‘batch mode’ for running simulations. This allows the user to superimpose traces from multiple simulations. In the batch simulation (i.e., trt.bch), the first simulation contains only the excitatory input. The second simulation contains both inhibitory and excitatory inputs. The results of this batch simulation are illustrated in Segev_bch.gif. This results can be compared to Fig. 3.4 of Segev et al. 1989.