Conformation Stability Assay for SAA Amplified Materials
Stability Procedure
- In ultracentrifuge tubes (230 uL capacity), incubate sample with 0, 1, 2, 3, 4, 5, and 6 M GdnHCl at room temperature for 2 hours. Cover with parafilm and vortex gently every 30 minutes.
| Tube No. | Final [GdnHCl] (M) | Denaturant Volume (μL) | Denaturant | dH2O (μL) | SAA Sample Volume (μL) |
| 1 | 0.0 | 10 | PBS (1x) | NA | 10 |
| 2 | 1.0 | 10 | 2 M GdnHCl | NA | 10 |
| 3 | 2.0 | 10 | 4 M GdnHCl | NA | 10 |
| 4 | 3.0 | 10 | 6 M GdnHCl | NA | 10 |
| 5 | 4.0 | 10 | 8 M GdnHCl | NA | 10 |
| 6 | 5.0 | 12.5 | 8 M GdnHCl | 2.5 | 5 |
| 7 | 6.0 | 15 | 8 M GdnHCl | NA | 5 |
- Following incubation, normalize the concentration of GdnHCl in all samples to 0.55 M by adding the following volumes of PBS (1x) and 8 M GdnHCl. Pipette mix slowly.
Note: Add 8 M GdnHCl last to prevent unwanted denaturation of lower concentration samples.
| [GdnHCl] (M) | 8 M GdnHCl (μL) | PBS (1x) (μL) |
| 0 | 15.13 | 184.87 |
| 1 | 12.65 | 187.35 |
| 2 | 10.18 | 189.82 |
| 3 | 7.70 | 192.30 |
| 4 | 5.23 | 194.77 |
| 5 | 2.75 | 197.25 |
| 6 | 0 | 200.00 |
- Centrifuge samples with a Beckman 42.2 Ti rotor at 100,000 x g at 4oC for 1 hour.
- Remove supernatant.
- Resuspend the pellet by adding 150 μL of NuPage (2x) loading buffer to each sample, and heating in a 95oC water bath for 10 minutes.
- Run SDS-PAGE gel using 12% Bis-tris gels with MES (1x) running buffer at 155 V for 75 minutes.
Note: Load 2x volume for 5 M and 6 M GdnHCl samples.
- Transfer protein to the nitrocellulose membrane for 1 hour at 100 V.
- Block the membrane with 5% milk for 1 hour.
- Incubate the membrane with anti-aSyn antibody (BD clone 42; 1:1000) overnight at 4o
- Wash the membrane three times, 10 minutes each, with PBS-T.
- Incubate the membrane with anti-mouse secondary antibody (Sigma; 1:2000) for 1 hour at room temperature.
- Wash the membrane three times, 10 minutes each, with PBS-T.
- Develop with ECL and image.
Western Blot Analysis and Denaturation Curve Procedure
- Open WB JPG file in ImageJ.
- Subtract background from image.
a. Process -> Subtract Background -> Set ‘Rolling ball radius:’ to 50.0 pixels* -> Select ‘Light background’ -> OK (*Set rolling ball radius higher for less subtraction)
- Use the Rectangle tool to select the first lane (0 M GdnHCl). Press ‘1’ on keyboard to label as the first lane. Reposition selection box to each consecutive lane and press ‘2’ to label each one. Press ‘3’ to label the last lane. ImageJ will automatically graph the density of the bands in each well.
- Use the Line tool to enclose the bottom of each curve.
- Use the Wand tool to select the area under each curve. A window will open with the area.
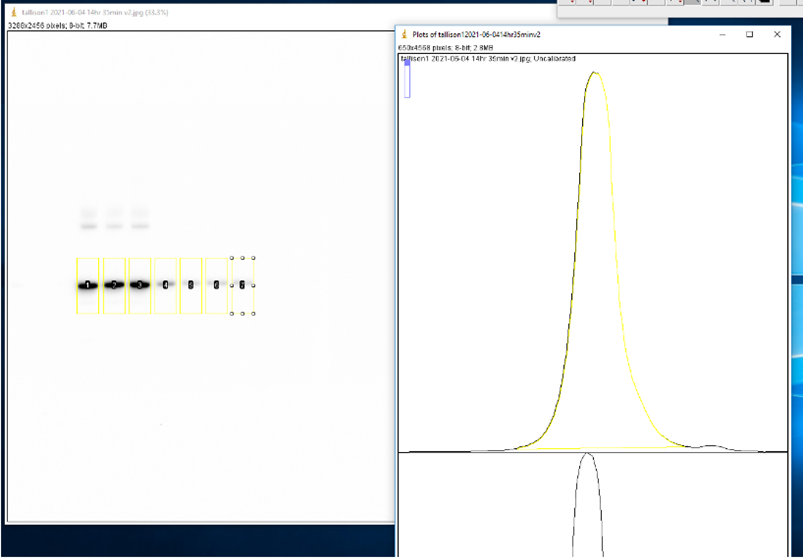
- Copy the area values to an XY table in GraphPad Prism version 9.1.1 with a single Y value for each point.
- Normalize the density values.
a. Analyze -> Data Processing -> Normalize -> Define 0% as Y=0, and 100% as “Last value in each data set (or first, whichever is larger) -> OK
- A new results table will be created with the normalized values.
- Create a Nonlinear Regression Curve to determine the GdnHCl50 (the guanidine concentration at with half of the aggregates have been denatured).
a. Analyze -> Regression and Curves -> Nonlinear Regression (Curve Fit) -> OK -> Select ‘Sigmoidal dose-response (variable slope)’ -> New -> Clone the selected equation -> Select ‘Default Constraints’ tab -> Change the bottom parameter to ‘Constant equal to 0’ and the top parameter to ‘Constant equal to 100’ -> OK -> Select the new equation created -> OK
b. A new results tab will be created with the non-linear fit analysis. The ‘LogEC50’ values will be the GdnHCl50.